chlamydia nucleic acid amplification tests
- 2. (NAATs) supervisor:Dr Mohammad Reza Pourmand by: Morteza karami zarandi Chlamydia Nucleic Acids Amplification Tests
- 3. Chlamydia trachomatis • Obligate intracellular gram negative organism • Commonest bacterial STD • Disease due to direct damage to cells and immunopathology causing fibrosis and scarring • Diagnostic Options for Chlamydial Infections: – Cell Culture – Direct Fluorescence Assay – Enzyme Immunoassay – Hybridisation Assay – Nucleic Acid Amplification Tests
- 4. Nucleic Acid Amplification Tests (NAATs) • Present advantages – The most sensitive and specific assays – Self collected samples (vaginal and male urine) – Pooling specimens – Automation possible
- 5. NAATs • Present disadvantages – False positive result : contamination by amplicon – False negative result : inhibitors – Mutation target (swedish variant) – In low prevalence population, need to consider confirmation of positives
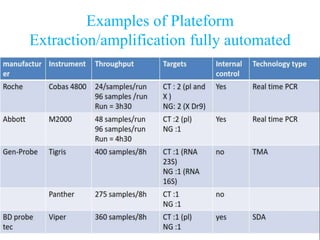
- 6. QCMD1 CT 2012 Details of commercial tests • PCR – Cobas Amplicor • Real time PCR – Roche cobas 4800 CT/NG test – Roche Cobas TaqMan CT test – Abbott Real time CT or CT/NG • TMA – Gen-Probe APTIMA Combo 2 or CT assay • SDA – BD Probe Tec – Viper Probe tec 1.Quality Control for Molecular Diagnostics
- 7. Examples of Plateform Extraction/amplification fully automated
- 8. Polymerase Chain Reaction(PCR) • The COBAS AMPLICOR system automates amplification and detection of target nucleic acids, making diagnostic PCR routine for a variety of infectious diseases. • The system contains – Single thermal cycler – Incubator – Magnetic particle washer – Pipettor – Photometer
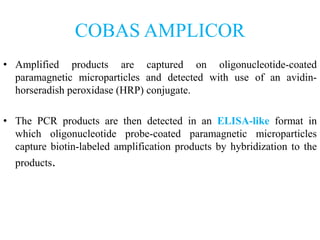
- 9. COBAS AMPLICOR • Amplified products are captured on oligonucleotide-coated paramagnetic microparticles and detected with use of an avidin- horseradish peroxidase (HRP) conjugate. • The PCR products are then detected in an ELISA-like format in which oligonucleotide probe-coated paramagnetic microparticles capture biotin-labeled amplification products by hybridization to the products.
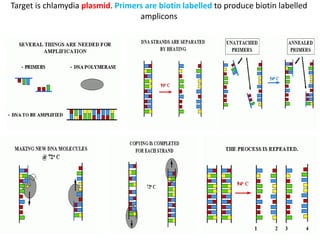
- 10. Target is chlamydia plasmid. Primers are biotin labelled to produce biotin labelled amplicons
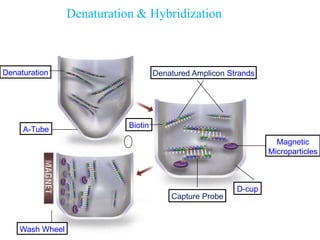
- 11. Denaturation & Hybridization Denaturation Denatured Amplicon Strands Biotin Magnetic Microparticles Capture Probe D-cup A-Tube Wash Wheel
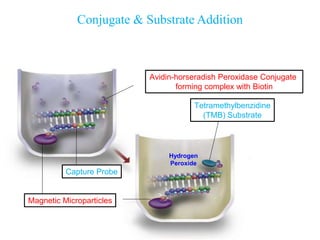
- 12. Conjugate & Substrate Addition • Avidin-horseradish Peroxidase Conjugate forming complex with Biotin Tetramethylbenzidine (TMB) Substrate Hydrogen Peroxide Magnetic Microparticles Capture Probe
- 13. samples • Urine Store up to 24h at RT and up to 7 days at 2 – 8 o c • Vaginal swab In amplicor specimen transport medium store at RT for up to 10 days.
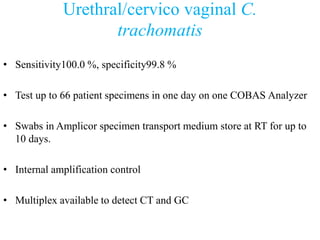
- 14. Urethral/cervico vaginal C. trachomatis • Sensitivity100.0 %, specificity99.8 % • Test up to 66 patient specimens in one day on one COBAS Analyzer • Swabs in Amplicor specimen transport medium store at RT for up to 10 days. • Internal amplification control • Multiplex available to detect CT and GC
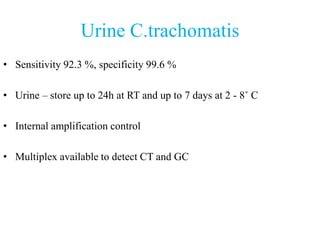
- 15. Urine C.trachomatis • Sensitivity 92.3 %, specificity 99.6 % • Urine – store up to 24h at RT and up to 7 days at 2 - 8˚ C • Internal amplification control • Multiplex available to detect CT and GC
- 16. Cobas Amplicor disadvantages • Positive internal control signal might be misleading in inhibitory specimens with low amount of C. trachomatis. • Roche method had difficulty in detecting dual infections, generally with male urine specimens, when there were relatively large amounts of Neisseria gonorrhoeae and small amounts of C. trachomatis target based on DC analysis
- 17. RealTime PCR • Commercial RealTime PCR tests: – Roche cobas 4800 – Abbott M 2000 CT/NG assay
- 18. Real Time PCR
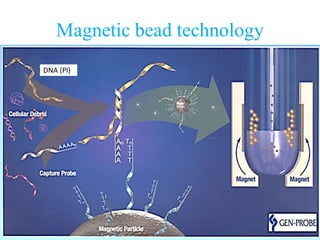
- 19. Abbott Real Time CT/NG assay • The Abbott method uses an automated nucleic acid extraction platform based on magnetic bead technology and robotic PCR setup, followed by the manual transfer of PCR mixtures to an m2000rt thermocycler for real-time PCR amplification and reading • Target is chlamydial plasmid
- 20. Magnetic bead technology DNADNA (Pl)
- 21. Abbott Real Time CT/NG assay • “CT” stands for Chlamydia trachomatis and “NG” stands for N.gonorrhoeae that uses the automated m2000 molecular platform • Abbot assay was more sensitive than the Roche assay for both C. trachomatis and N. gonorrhoeae. • Abbott RealTime CT/NG assay is an accurate and automated new addition to the available testing options for C. trachomatis and N. gonorrhoeae
- 22. Transcription-Mediated Amplification (TMA) • RNA transcription amplification system using two enzymes, reverse transcriptase and T7 RNA polymerase • Isothermal, amplification of rRNA target • Produces over ten billion-fold amplification • Single-tube format • Gen-Probe Aptima Combo 2 assay
- 23. APTIMA COMBO 2 ASSAY • It is a second generation nucleic acid amplification test that is used for in vitro detection & differentiation of rRNA from CT & GC. • AC2 assay is a qualitative molecular diagnostic assay • Since it provides qualitative results, there is no correlation between the magnitude of a positive assay signal & the number of organisms in a specimen.
- 24. APTIMA COMBO 2 ASSAY • Uses three technologies: – Target capture specimen processing (TCT) – Transcription-Mediated Amplification (TMA) – Dual Kinetic Assay detection (DKA)
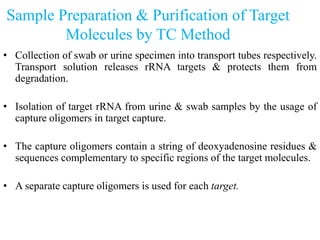
- 25. Sample Preparation & Purification of Target Molecules by TC Method • Collection of swab or urine specimen into transport tubes respectively. Transport solution releases rRNA targets & protects them from degradation. • Isolation of target rRNA from urine & swab samples by the usage of capture oligomers in target capture. • The capture oligomers contain a string of deoxyadenosine residues & sequences complementary to specific regions of the target molecules. • A separate capture oligomers is used for each target.
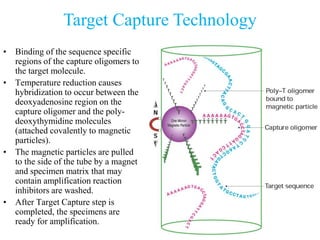
- 26. Target Capture Technology • Binding of the sequence specific regions of the capture oligomers to the target molecule. • Temperature reduction causes hybridization to occur between the deoxyadenosine region on the capture oligomer and the poly- deoxythymidine molecules (attached covalently to magnetic particles). • The magnetic particles are pulled to the side of the tube by a magnet and specimen matrix that may contain amplification reaction inhibitors are washed. • After Target Capture step is completed, the specimens are ready for amplification.
- 27. Advantages of Target Capture • Eliminates centrifugation and chemical extraction steps. • Allows simultaneous targeting of multiple nucleic acid sequences. • Allows the use of large volumes and a variety of samples that are currently difficult to amplify such as cerebral spinal fluid and serum. • Increases specificity, because target nucleic acids are purified before amplification. • Increases sensitivity by reducing the inhibition rate.
- 28. Transcription-Mediated Amplification • One primer contains a T7 promoter sequence for RNA polymerase that hybridizes to the target RNA • RT creates a cDNA of the target RNA by extension from the 3’ end of the promoter-primer • The RNA of the resulting RNA:DNA duplex is degraded by the RNAse H activities of the RT • A second primer then binds to the cDNA containing the promoter sequence from the T7 promoter- primer
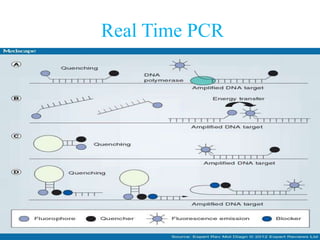
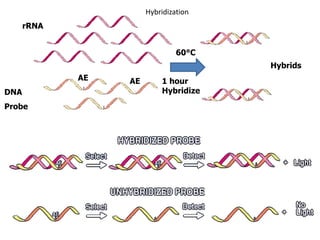
- 29. Amplicon Detection by Hybridization Protection Assay (HPA) • Single-stranded chemiluminescent DNA probes, are labeled with different acridinium ester molecules and are complementary to a region of each target amplicon. • Stable RNA:DNA hybrids are then formed from the labeled DNA probes binding to amplicon. • The Selection Reagent eliminates the generation of signal from unhybridized probe by differentiating hybridized from unhybridized probe. • In the detection step, light emitted from the labeled RNA:DNA hybrids is measured as photon signals and are reported as Relative Light Units (RLU) in a luminometer
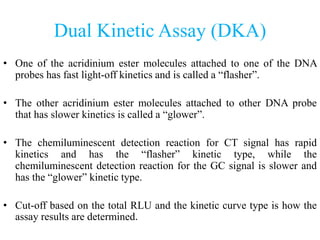
- 31. Dual Kinetic Assay (DKA) • One of the acridinium ester molecules attached to one of the DNA probes has fast light-off kinetics and is called a “flasher”. • The other acridinium ester molecules attached to other DNA probe that has slower kinetics is called a “glower”. • The chemiluminescent detection reaction for CT signal has rapid kinetics and has the “flasher” kinetic type, while the chemiluminescent detection reaction for the GC signal is slower and has the “glower” kinetic type. • Cut-off based on the total RLU and the kinetic curve type is how the assay results are determined.
- 32. Limitation of APTIMA Combo 2 Assay: • Therapeutic failure or success cannot be determined , since nucleic acid may persist after antimicrobial therapy. • It is not a quantitative assay. • This assay does not permit microscopic assessment of specimen adequacy, so training of the techniques is necessary.
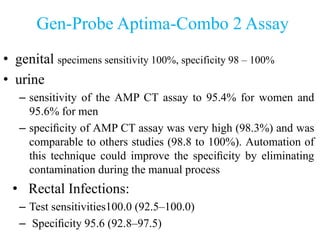
- 33. Gen-Probe Aptima-Combo 2 Assay • genital specimens sensitivity 100%, specificity 98 – 100% • urine – sensitivity of the AMP CT assay to 95.4% for women and 95.6% for men – specificity of AMP CT assay was very high (98.3%) and was comparable to others studies (98.8 to 100%). Automation of this technique could improve the specificity by eliminating contamination during the manual process • Rectal Infections: – Test sensitivities100.0 (92.5–100.0) – Specificity 95.6 (92.8–97.5)
- 34. Strand Displacement Amplification (SDA) • Target gene – Chlamydia plasmid • Target generation phase – dsDNA heat denatured to give ssDNA • Amplification phase – SDA isothermal. ssDNA amplified using DNA polymerase, restriction enzyme, primers with restriction enzyme recognition sites, bumpers • Detection phase – ET fluorescence energy transfer
- 35. Becton Dickinson ProbeTec SDA ssDNA with restriction site Amplification primer binds DNA polymerase extends primer dsDNA with restriction sites
- 36. BD ProbeTec SDA Restriction enzyme binds Nicks one strand dsDNA DNA polymerase binds, extends displacing previously made strand Restriction site repeatedly nicked and new strands made and displaced Each displaced strand enters cycle DNA strands bind to probe and detected by Energy Transfer Hairpin secondary structure Fluorescent dye Quenching dye
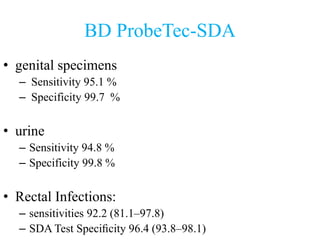
- 37. BD ProbeTec-SDA • genital specimens – Sensitivity 95.1 % – Specificity 99.7 % • urine – Sensitivity 94.8 % – Specificity 99.8 % • Rectal Infections: – sensitivities 92.2 (81.1–97.8) – SDA Test Specificity 96.4 (93.8–98.1)
- 38. Ideal Test • High sensitivity and specificity • Suitable for non invasive samples – urine • Fast turnaround time • Low cost • Not affected by inhibitory substances • Transport and storage of samples not critical • Able to automate
- 39. Ideal Test • Gen-Probe assays detected all mutant strains characterized by the in- house PCR as having the deletion in the cryptic plasmid, whereas the Roche and the Abbott PCRs targeting the plasmid were both unable to detect the plasmid mutant. • Gen-Probe assays can perform 400 sample in 8 hourse • Gen-Probe assays Eliminates centrifugation and chemical extraction steps
- 40. References 1. Møller JK1, Pedersen LN, Persson K. Comparison of Gen-Probe Transcription-Mediated Amplification, Abbott PCR, and Roche PCR Assays for Detection of Wild-Type and Mutant Plasmid Strains of Chlamydia trachomatis in Sweden. J Clin Microbiol. 2008 Dec;46(12):3892-5. doi: 10.1128/JCM.00412-08. Epub 2008 Oct 8. 2. Møller JK1, Pedersen LN, Persson K. Comparison of the Abbott RealTime CT New Formulation Assay with Two Other Commercial Assays for Detection of Wild-Type and New Variant Strains of Chlamydia trachomatis .J Clin Microbiol. 2010 Feb;48(2):440-3. doi: 10.1128/JCM.01446-09. Epub 2009 Dec 9 3. DiDomenico N1, Link H, Knobel R, et al. COBAS AMPLICORTM: fully automated RNA and DNA amplification and detection system for routine diagnostic PCR . Clin Chem. 1996 Dec;42(12):1915-23. 4. de Barbeyrac B1, Géniaux M, Hocké C,et al. Detection of Chlamydia trachomatis in symptomatic and asymptomatic populations with urogenital specimens by AMP CT (Gen- Probe Incorporated) compared to others commercially available amplification assays. Diagn Microbiol Infect Dis. 2000 Jul;37(3):181-5
- 41. 5. Akduman D1, Ehret JM, Messina K, et al. Evaluation of a Strand Displacement Amplification Assay (BD ProbeTec-SDA) for Detection of Neisseria gonorrhoeae in Urine Specimens .J Clin Microbiol. 2002 Jan;40(1):281-3. 6. Schuurs TA, Verweij SP, Weel JFL, et al. Detection of Chlamydia trachomatis and Neisseria gonorrhoeae in an STI population: performances of the Presto CT-NG assay, the Lightmix Kit 480 HT CT/NG and the COBAS Amplicor with urine specimens and urethral/ cervicovaginal samples. BMJ Open 2013;3:e003607. doi:10.1136/bmjopen-2013- 003607 7. Laura H. Bachmann, Robert E. Nucleic Acid Amplification Tests for Diagnosis of Neisseria gonorrhoeae and Chlamydia trachomatis Rectal Infections. J Clin Microbiol. May 2010, p. 1827–1832 Vol. 48, No. 5 0095-1137/10/$12.00 doi:10.1128/JCM.02398-09 8. Annie Cheng, Qinfang Qian, and James E. Kirby Evaluation of the Abbott RealTime CT/NG Assay in Comparison to the Roche Cobas Amplicor CT/NG Assay JOURNAL OF CLINICAL MICROBIOLOGY, Apr. 2011, p. 1294–1300 Vol. 49, No. 4 0095- 1137/11/$12.00 doi:10.1128/JCM.02595-10 9. M. Chernesky,a D. Jang,a E. Portillo,a M. Smieja,a J. Kapala,b C. Doucette,b J. Sumner,b R. Ewert,c C. MacRitchie,d and J. Gilchrista Comparison of Three Assays for Detection of Chlamydia trachomatis and Neisseria gonorrhoeae in SurePath Pap Samples and the Role of Pre- and Postcytology Testing
- 42. 10. Fadiman, K., Goldman, S. (2004). Screening for Chlamydia trachomatis American College of Preventive Medicine. San Francisco, CA. pp. 1-3 11. Gen-Probe (2000). New Directions in Molecular Diagnostic Testing Pioneer advances in Health Care. CA.pp3-12