Abstract
Free full text

Etoposide pathway
Description
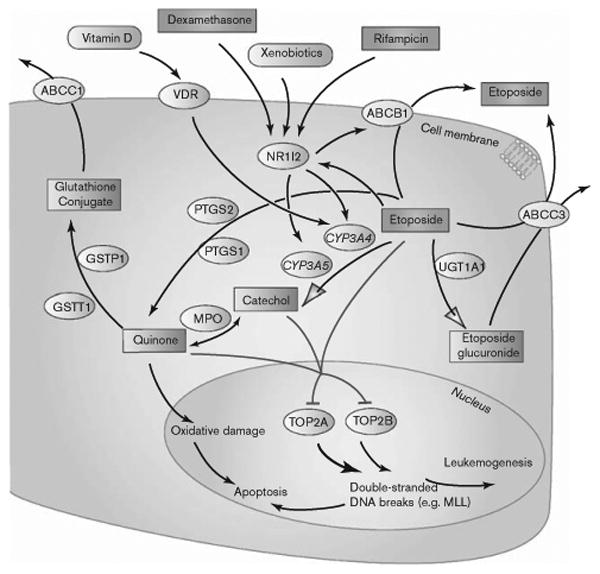
Etoposide is a commonly used chemotherapy agent with a broad range of antitumor activity. Etoposide and teniposide, the epipodophyllotoxins, stabilize the double-stranded DNA cleavage normally catalyzed by topoisomerase II (topo II) and inhibit faithful religation of DNA breaks [1,2]. These double-strand DNA breaks subsequently trigger the desired antitumor effects of the drugs. Metabolism of etoposide is mediated by CYP3A4 and CPY3A5 (Fig. 1) [3,4], both of which are transcriptionally regulated by NR1I2 (i.e. pregnane X receptor). Thus, xenobiotics that modulate NR1I2 activity (e.g. dexamethasone and rifampicin) have been observed to enhance etoposide clearance [5,6]. In addition to CYP3A4/5 mediated reactions, conversion of etoposide to the O-demethylated metabolites (catechol and quinone) can also be catalyzed by prostaglandin synthases or myeloperoxidase [7–9]. These metabolites have similar potency at inhibiting topoisomerase II and are more oxidatively reactive than the parent drug [10]. Glutathione [11] and glucuronide conjugation [12] seem to inactivate parent drug and metabolite, and are mediated by GSTT1/GSTP1 and UGT1A1 [13,14], respectively. Efflux of conjugated or unconjugated forms of etoposide has been associated with ABCC1, ABCC3, and ABCB1 [15,16], representing plausible mechanisms of drug resistance. Epipodophyllotoxins are highly effective anticancer agents, but can cause a delayed toxicity: treatment-related acute myeloid leukemia or myelodysplastic syndrome [17–19]. Drug-induced formation of MLL fusion genes has been associated with the development of treatment-related acute myeloid leukemia or myelodysplastic syndrome [20]. Although etoposide inhibits both topo II alpha and beta, the antitumor activity of etoposide is shown to be delivered primarily through inhibition of topo II alpha [21], whereas the carcinogenic effect has been attributed to the beta isoform [22]. Recently, 64 genetic variants that contribute to etoposide-induced cytotoxicity were identified through a whole-genome association study [23].
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website (http://www.pharmgkb.org/do/serve?objId=PA2025&objCls=Pathway).
References
Full text links
Read article at publisher's site: https://doi.org/10.1097/fpc.0b013e32832e0e7f
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc4164627?pdf=render
Subscription required at www.jpharmacogenetics.com
http://content.wkhealth.com/linkback/openurl?issn=1744-6872&volume=19&issue=7&spage=552
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1097/fpc.0b013e32832e0e7f
Article citations
Hemophagocytic Lymphohistiocytosis Activation Syndrome with Multi-Organ Co-Infections: A Therapeutic Dilemma.
Kans J Med, 16:268-270, 30 Oct 2023
Cited by: 0 articles | PMID: 37954881 | PMCID: PMC10635691
Isolation of circulating endothelial cells provides tool to determine endothelial cell senescence in blood samples.
Sci Rep, 14(1):4271, 21 Feb 2024
Cited by: 2 articles | PMID: 38383692 | PMCID: PMC10882010
Signatures of Co-Deregulated Genes and Their Transcriptional Regulators in Kidney Cancers.
Int J Mol Sci, 24(7):6577, 31 Mar 2023
Cited by: 2 articles | PMID: 37047552 | PMCID: PMC10094846
Etoposide, an anticancer drug involved in therapy-related secondary leukemia: Enzymes at play.
Transl Oncol, 14(10):101169, 06 Jul 2021
Cited by: 23 articles | PMID: 34243013 | PMCID: PMC8273223
Review Free full text in Europe PMC
Sensitization of the UPR by loss of PPP1R15A promotes fibrosis and senescence in IPF.
Sci Rep, 11(1):21584, 03 Nov 2021
Cited by: 7 articles | PMID: 34732748 | PMCID: PMC8566588
Go to all (26) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Modulation of drug sensitivity in yeast cells by the ATP-binding domain of human DNA topoisomerase IIalpha.
Nucleic Acids Res, 31(19):5714-5722, 01 Oct 2003
Cited by: 8 articles | PMID: 14500835 | PMCID: PMC206448
Resveratrol induces DNA double-strand breaks through human topoisomerase II interaction.
Cancer Lett, 295(2):167-172, 20 Mar 2010
Cited by: 28 articles | PMID: 20304553
Analysis of etoposide binding to subdomains of human DNA topoisomerase II alpha in the absence of DNA.
Biochemistry, 40(6):1624-1634, 01 Feb 2001
Cited by: 33 articles | PMID: 11327821
[DNA topoisomerases targeting anticancer agents and mechanism for acquirement of drug resistance].
Nihon Rinsho, 55(5):1096-1102, 01 May 1997
Cited by: 0 articles | PMID: 9155159
Review