March 13, 2023 report
This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Single-cell sequencing solution seeks to unleash disruptive science, with a vortexer
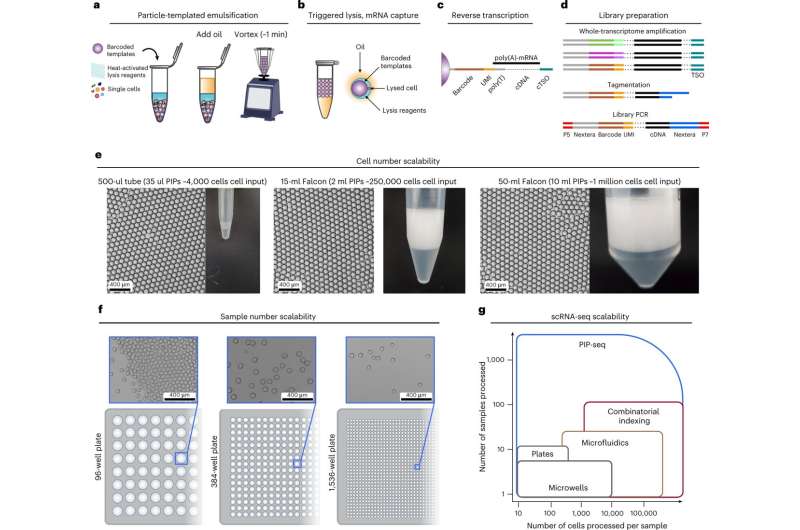
A new single-cell encapsulation, lysis and barcoding method of cDNA is faster and requires less equipment, hardware, expense and expertise. It has been demonstrated to be compatible and scalable with any size container, from 500uL microcentrifuge tubes to 50 ml conical tubes, and works with 96-, 384- or 1,536-well microtiter plates, as well.
In their paper, "Microfluidics-free single-cell genomics with templated emulsification," published in Nature Biotechnology, the researchers estimate that 3,500 cells can be barcoded with 35 µl of hydrogel templates in a 500-µl tube, 225,000 cells with 2 ml of templates in a 15-ml conical tube, and 1 million cells with 10 ml of templates in a 50-ml tube. Regardless of the tube size, only 2 min of vortexing is required for cell capture.
Particle-templated instant partition sequencing (PIP-seq) captures cells, barcoded templates, and lysis reagents in uniform oil-coated water droplets with only a few minutes of vortexing. The process uses hydrogel particles constituting a >95% water solution as templating to obtain these well-defined droplets. The hydrogels with pre-functionalized beads are filled by single cells and heat-activated lysing solution during vortexing.
The cells are then lysed by increasing the temperature to 65 °C, which activates a proteolytic enzyme (proteinase K) that breaks down the cell plasma membrane, releasing the cellular mRNA. The mRNA is then captured on polyacrylamide beads decorated with barcoded sequences.
PIP-seq emulsions can be stored at 0 °C without any change in data quality (72 hours), allowing multiple samples to be stacked for sequencing. After resuming, oil is removed, beads are transferred into a reverse transcription buffer, and full-length cDNA is synthesized, amplified, and prepared for sequencing.
Single-cell transcriptomics is gaining an increasingly high level of focus in research as it allows scientists to understand in great detail what is happening within a cell. Applications range from categorizing active cell functions to determining disease cause or progression to uncovering hidden RNA regulators and specific interactions, much of which can only be crudely inferred in bulk-cell studies.
In the paper, the researchers demonstrate that PIP-seq produces high-purity transcriptomes in a mouse–human cell mixing test. Looking for the degree to which pre-lysed cells could result in mRNA cross-contamination. The trial found the fraction of mouse reads in human transcriptomes was below 3%.
PIP-seq was also tested in single-cell transcriptional profiling of mixed phenotype acute leukemia. Here the researchers identified transcriptional differences beyond immunophenotype observation. The modulation of ribosomal genes, previously unconnected to this type of leukemia, could play a role in treatment resistance and suggests a therapeutic target to the researchers—an impressive find for a method testing study that illustrates a blindspot of other methods.
Disruptive innovation
When scientists talk about disruptive innovations or technology, they generally refer to a new method that changes how research is pursued. Typically this means making an existing process cheaper, faster, more accurate, more precise or at an increased throughput of data collection. Much like how cell phones replaced landlines, online streaming replaced video stores, or next-generation sequencing replaced Sanger sequencing, disruptive innovations are often quickly adopted and increase the quality and rate of research work.
PIP-seq emulsion capture is equivalent to what can be achieved with microfluidic systems but at a fraction of the cost with no major equipment investment, making it a simple, flexible, and scalable next-generation sequencing workflow that extends single-cell sequencing to new applications—making it a truly disruptive innovation in single-cell research. The PIP-seq inventors hope this process will open up the field of single-cell sequencing to smaller, less-resourced research organizations and academic institutions so they can be free to pursue the next wave of scientific discoveries.
More information: Iain C. Clark et al, Microfluidics-free single-cell genomics with templated emulsification, Nature Biotechnology (2023). DOI: 10.1038/s41587-023-01685-z
Journal information: Nature Biotechnology
© 2023 Science X Network





















