Abstract
The proteins encoded by the classical HLA class I and class II genes in the major histocompatibility complex (MHC) are highly polymorphic and play an essential role in self/non-self immune recognition. HLA variation is a crucial determinant of transplant rejection and susceptibility to a large number of infectious and autoimmune disease1. Yet identification of causal variants is problematic due to linkage disequilibrium (LD) that extends across multiple HLA and non-HLA genes in the MHC2,3. We therefore set out to characterize the LD patterns between the highly polymorphic HLA genes and background variation by typing the classical HLA genes and >7,500 common single nucleotide polymorphisms (SNPs) and deletion/insertion polymorphisms (DIPs) across four population samples. The analysis provides informative tag SNPs that capture some of the variation in the MHC region and that could be used in initial disease association studies, and provides new insight into the evolutionary dynamics and ancestral origins of the HLA loci and their haplotypes.
Numerous studies have demonstrated association between HLA alleles and disease susceptibility (a partial list is provided in Table 1 and Supplementary Table 1), but the interpretation of these results is confounded by the strong correlation between alleles at neighboring HLA and non-HLA genes. Major efforts have therefore been directed at cataloguing the gene and variation content of the entire MHC4-6. In addition, previous studies in European-derived populations have examined the distribution of LD across the region and have suggested that SNPs could help dissect causal variation within the MHC2,3,7-10. Here, we have created a resource to guide future association studies by genotyping genetic variants across the extended MHC region of 7.5 Mb at a higher density and in more DNA samples than previously reported. In 361 individuals of African (YRI), European (CEU), Chinese (CHB), and Japanese (JPT) ancestry, the inferred haplotype structure across the region shows that LD is systematically higher in CEU, CHB and JPT samples than in the YRI sample (Fig. 1). Alleles across the different classical HLA loci demonstrate strong correlation (Supplementary Table 2). These high levels of LD among SNPs and DIPs and among HLA alleles suggest that SNPs outside the HLA genes are informative about HLA types (Fig. 2a), and that a few, well chosen SNPs may capture common classical HLA variation at several loci.
Table 1. Examples of HLA alleles associated with common disease and their tag SNPs.
| Phenotypea | Risk allele | Estimated relative risk | Tag SNPsb | r 2 c |
|---|---|---|---|---|
| Graves’ | B*0801/DRB1*0301/DQA1*0501/DQB1*0201 | 4 / 2.5 | rs3129763(C)+rs4639334(C) | 0.96 |
| disease / Myasthenia gravis | ||||
| Multiple sclerosis | DRB1*1501/DQB1*0602 | 4 | rs3135388 | 0.97 |
| Multiple sclerosis | DQA1*0102 | 4 | rs9268428(C)+rs6457594(A)+rs7451962(C) | 0.90 |
| Psoriasis | C*0602 | 5 | rs887466(G)+rs4379333(C) | 1.0 |
| Celiac diseased | DQA1*0201-DQB1*0202 (DQ2.2) DQA1*0501-DQB1*0201 (DQ2.5) |
1 7 |
rs4988889(T)+rs2858331(C) rs4988889(T)+rs2858331(T) |
1.0 0.93 |
| Systemic lupus erythematosus | DRB1*1501 | 2 | rs3135388 | 0.97 |
| Type 1 diabetes / SLE | DRB1*0301 | 4.5 | rs2040410 | 0.87 |
| Abacavir Hypersensitivity | B*5701 | 4 | rs2395029 | 1.0 |
A more complete list of disease associated alleles can be found in Supplementary Table 1, and tags for the HLA alleles in Supplementary Table 3.
Tags picked from CEU samples to capture the HLA risk allele. For multi-marker (haplotype) tests, alleles of the individual SNPs are also listed in parentheses. For many HLA alleles, there are likely to exist multiple equivalent tags/tests.
Coefficient of determination (r2) between the tag/test and the HLA risk allele in the CEU panel.
DQA1*0201/DQB1*0202 (DQ2.2) has no effect on its own. Only when a person carries the DQ2.2 in combination with DQA1*0501/DQB1*0201 (DQ2.5) does it increase risk. The relative risk of DQ2.5 changes depending on the specific combination with other DQ types.
Figure 1.
The relationship between recombination rates and haplotype structure spanning the 7.5 Mb extended MHC region (defined by the SLC17A2 gene at the telomeric end to the DAXX gene at the centromeric end of chromosome 6). Recombination rates (blue lines in cM/Mb) were estimated separately from each population and combined to provide a single estimate for the region. Recombination hotspots with strong statistical support in all three (in red) or two analysis panels (in pink) are indicated by the red triangles and vertical grey lines. The average recombination rate across the region is 0.44 cM/Mb, compared to a genome-wide average of 1.2 cM/Mb, and is particularly low in the 3 Mb region that includes the olfactory receptor gene cluster which has only two hotspots with strong statistical evidence33. The horizontal lines indicate the extent of non-redundant haplotypes (see text for details) identified in each analysis panel (YRI: green, CEU: orange and CHB+JPT: purple). Haplotypes are typically longer in regions of low recombination, and are often, though not always, interrupted by recombination hotspots. Haplotypes are typically longer in the CEU and CHB+JPT analysis panels than in YRI. The physical locations of the six classical HLA loci analyzed in this study are also shown.
Figure 2.
Allelic association between SNPs across the 7.5 Mb extended MHC region and HLA types at each gene for the combined population data using the 5,754 SNPs that were typed in all populations and are polymorphic across the combined population samples (see Methods for details). (a) The extent of association between SNPs across the region and HLA types at HLA-A (red), HLA-C (light green), HLA-B (dark green), HLA-DRB (blue), HLA-DQA (violet), HLA-DQB (purple) as measured by relative information in the combined population data. The significant information contained within these SNPs located outside the HLA genes is not surprising given the extensive LD between SNPs and HLA loci. LD extended up to 1 Mb from the centre of a given HLA gene and, as a consequence, a single SNP could be informative for more than a single HLA gene. (b) For HLA-C (the position of which is indicated by the vertical blue line), the position of SNPs across the 7.5 Mb region showing weak (0.2 < r2 <0.5; grey), moderate (0.5 < r2 < 0.8; blue) and strong (r2 > 0.8; red) association to each type that is present in each of the four populations. The size of the adjacent green bar indicates the relative frequency of each type in each population (types not present in a population are not shown).
We examined the association between HLA types and single SNPs across the entire region. Fig. 2b shows the results for HLA-C (see Supplementary Fig. 1 for the other HLA genes). In the four populations studied, 34-44% of the HLA alleles present are strongly associated with one or more individual SNPs (maximum r2 > 0.8), sometimes located at a considerable distance from the HLA allele. There are noticeable differences between the four populations studied. For example, allele HLA-C*0702 has many SNPs in moderate to strong LD in YRI and CEU extending over several Mb, while in CHB and JPT, strong association is only found to SNPs within 50 kb of the gene. In contrast, some alleles, such as HLA-C*0304, are not strongly associated with any single SNP in any of the four population samples studied in which it was found. These results suggest that while tagging of certain common HLA alleles in some populations may be relatively straightforward, tag SNPs are likely to differ between populations, and tagging of some HLA alleles may prove difficult if based solely on pairwise association to single SNPs.
To assess the extent to which allelic variation at HLA loci can be captured by nearby SNPs we used the “Tagger” algorithm11 to identify allelic tests using single SNPs or haplotypes of combinations of up to three SNPs as predictors of HLA. Following this tagging approach, the majority of common HLA alleles could be captured effectively and efficiently (Table 1 and Supplementary Table 3). We observed differences in the tagging performance between HLA genes: common (≥5%) alleles of HLA-A, -B, -C were captured, on average, with a maximum r2 = 0.97 in all four population samples, compared with a maximum r2 = 0.90 for all common HLA-DRB1 alleles. Of the less common (<5%) HLA alleles, 75% in YRI and CEU, and 100% in CHB and JPT are captured with a maximum r2 > 0.8, but one should exercise caution in interpreting these results given the small sample size and inaccuracies in allele frequency estimates.
The majority (~70%) of the HLA alleles are captured with high(er) r2 by specified haplotypes of multiple SNPs. Generally, a tag/test to capture a HLA allele observed in one reference panel captured that allele with lower r2 in the other population samples (Supplementary Table 4). This is broadly consistent with observed tag SNP transferability patterns in population samples across the major continents12. Additional empirical data in other samples is required to better understand the extent of transferability of tags selected across the MHC.
To this end, we performed an empirical validation of the tags for four different HLA alleles in two independent samples from ongoing disease studies. Specifically, we had access to 330 Dutch samples from a celiac disease study13 for which we had HLA typing data for DQA1 and DQB1 and 332 trio samples from a UK systemic lupus erythematosus (SLE) study14 for which we had HLA typing data for DRB1. The haplotype formed by the DQA1*0501 and DQB1*0201 alleles (also known as haplotype DQ2.5) is a known risk factor for celiac disease, with the highest risk for individuals homozygous for the DQ2.5 haplotype or that have one copy of this haplotype and one haplotype formed by DQA1*0201 and DQB1*0202 (haplotype DQ2.2)15. In SLE, significant association has been observed for both DRB1*1501 and DRB1*030116. We directly evaluated the predictive power of the SNPs/haplotypes for these alleles (Table 1) in these samples, and found that the sensitivity and specificity of these tags was significant and useful, not least, for example, in pre-screening large samples in the selection of certain individuals for further study (Table 2).
Table 2. Empirical validation of SNP-based tags of associated HLA alleles in large patient collections.
| HLA allele | Test based on SNP or haplotype | ||||||
|---|---|---|---|---|---|---|---|
| + | - | Total | Sensitivity (%) | Specificity (%) | r 2 | ||
| Celiac disease | |||||||
| DQA1*0201-DQB1*0202 (DQ2.2) / DQA1*0501- DQB1*0201 (DQ2.5) heterozygote |
+ - Total |
56 1 57 |
2 271 273 |
58 272 330 |
96.6 |
99.6 |
0.94 |
|
| |||||||
| DQA1*0501-DQB1*0201 (DQ2.5) homozygote |
+ - Total |
72 3 75 |
3 252 255 |
75 255 330 |
96.0 |
98.8 |
0.90 |
|
| |||||||
| SLE | |||||||
| DRB1*1501 | + - Total |
161 12 173 |
6 1149 1155 |
167 1161 1328 |
96.4 |
99.0 |
0.88 |
|
| |||||||
| DRB1*0301 | + - Total |
245 24 269 |
5 1054 1059 |
250 1078 1328 |
98.0 |
97.8 |
0.87 |
The test for DQ2.2 (DQA1*0201/DQB1*0202) is the rs4988889(T), rs2858331(C) haplotype
The test for DQ2.5 (DQA1*0501/DQB1*0201) is the rs4988889(T), rs2858331(T) haplotype
The test for DRB1*1501 is rs3135388; The test for DRB1*0301 is rs2187688
In general, two features make HLA allele tagging more difficult than tagging of SNPs. First, HLA alleles are themselves multilocus haplotypes, identified by unique combinations of sequence motifs generated by mutation, recombination and gene conversion17. Second, the unique evolutionary history of the MHC means that patterns of association are not just influenced by recombination, gene conversion, demography and genetic drift, but also through natural selection. In particular, HLA class I and class II alleles are often maintained in the population by balancing selection18-20 (e.g. heterozygote advantage, frequency-dependent selection). Certain forms of balancing selection, such as host-pathogen frequency-dependent selection21, will favor novel combinations of alleles across multiple HLA genes, hence actively selecting for recombinants20. However, as favored HLA combinations increase in frequency, so will the haplotype background on which they occurred. The direct consequence of such a dynamic is that a given HLA allele might be found on one, two, or several different haplotype backgrounds depending on where in the cycle of fluctuating selection it currently lies. In addition, balancing selection has resulted in the existence of hundreds of HLA alleles and haplotypes in populations, the vast majority of which are not common (less than 2% frequency), and yet collectively account for a significant proportion of the genetic variation. Given the limitations of this MHC variation resource (in terms of density and sample size), it remains to be seen how well the less common variants, haplotypes or recombinants can be captured via a tagging approach.
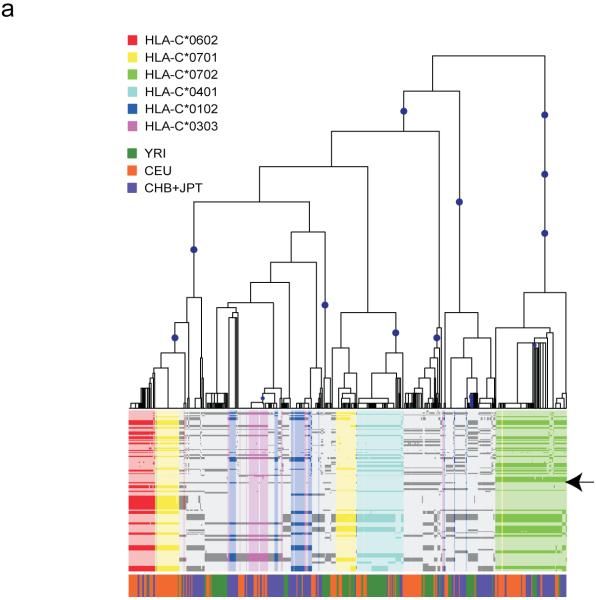
To illustrate the link between tagging efficiency and evolutionary dynamics we mapped the distribution of common alleles to the evolutionary tree relating haplotypes around the HLA-C gene (Fig. 3a). Certain common alleles, such as C*0702, are associated with a single clade in the tree that is defined by multiple SNPs. Such an allele can therefore be tagged with high efficiency, and its tags will likely be transferable between populations. The evolutionary implication is that this allele has a recent origin, or that a recent recombinant haplotype carrying this allele has been favored by natural selection coupled to the loss from all populations of the allele on any other haplotype background by random drift and bottleneck effects. In contrast, allele C*0701 occurs on two quite distinct clades of the tree that differ considerably in frequency between populations, an observation supported by analysis of long-range haplotype structure (Fig. 3b). Both of these clades are of recent origin (as indicated by their extensive haplotype backgrounds) such that they can be tagged, though only through combinations of multiple SNPs. Other alleles are yet further dispersed across the evolutionary tree and consequently harder to tag; for example, C*0303 requires two tags in CEU and CHB and three in JPT (it is absent from YRI), and no single tag SNP was selected in more than one population. Identification of differences in evolutionary history, for HLA alleles associated with disease, is informative for association mapping experiments, as specific HLA alleles distributed in different clades will carry different sets of linked HLA and non-HLA alleles. Identification of the causal allele(s) will depend on the ability to distinguish the clade associated with disease, followed by direct re-sequencing of the corresponding chromosomes to identify candidate variants.
Figure 3.
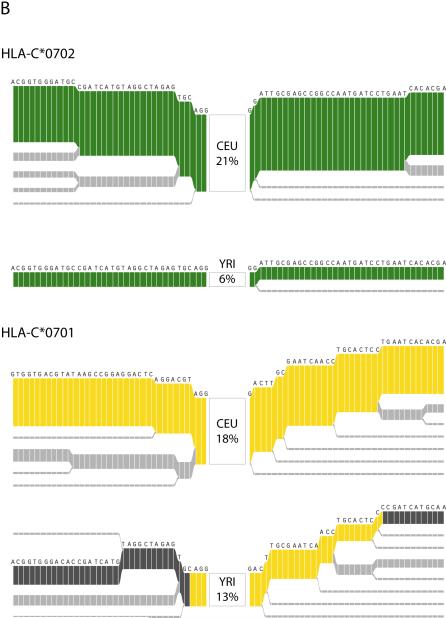
The evolutionary history of HLA-C. (a) Estimated evolutionary tree showing relationships among haplotypes at the HLA-C locus (defined as position 31,341,277 in build 34 or between SNPs rs2853950 and rs2001181) with mutations (blue circles) that unambiguously determine clades in the tree (see Methods for details). Below is a plot of the 478 haplotypes observed in the 100 kb region surrounding HLA-C (each column is a single haplotype) with the less common allele shown in the darker color. Colors indicate the HLA-C allele carried on each haplotype for the six most common alleles (each seen at least 30 times in the combined populations), with the position of HLA-C indicated by the arrow. The colored bar below indicates the population origin of each haplotype (YRI: green, CEU: orange and CHB+JPT: purple). Some alleles such as HLA-C*0702 (green) cluster within the tree whereas others such as HLA-C*0701 (yellow) occur in two or more parts of the tree. Furthermore, the two clades representing HLA-C*0701 are at different frequencies in the four populations. (b) Long-range haplotype structure around alleles C*0702 and C*0701. For C*0702, the common long-range haplotype is shared among the 2 populations, CEU and YRI, and is accordingly associated with a single clade. In contrast, for C*0701, the long-range haplotypes that are common in CEU and YRI are divergent. A shared haplotype structure nearby the HLA allele suggests that allele had a common origin in the 2 populations. A recombination event, however, likely occurred in at least one of the populations, placing them in 2 different clades.
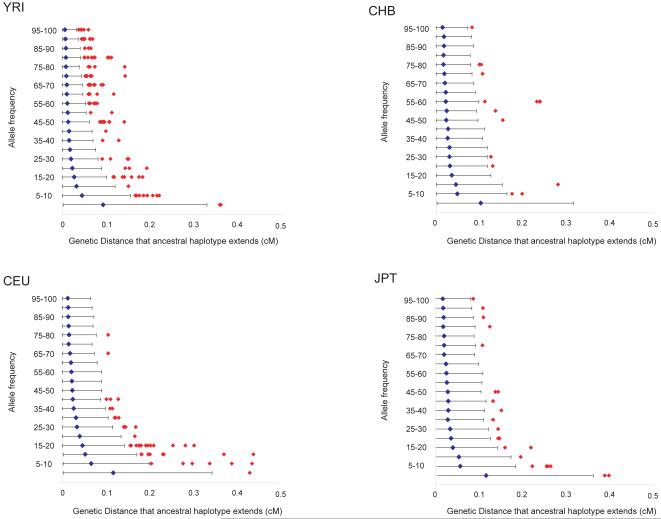
Identification of alleles in the MHC that have likely undergone positive selective pressure can also provide candidates to test for association to immune mediated diseases. Preliminary searches of the CEU population indeed suggested that such alleles were also associated with autoimmunity3. One approach for identifying recent positive selection is to identify alleles that are prevalent, but that are associated with long-range haplotypes, unbroken by recombination over time (suggesting they are of young age) 22. We used this ‘long-range haplotype’ approach on the current data set with the matched genome-wide data available from HapMap, resulting in the identification of several alleles within the MHC region that show evidence for recent selective sweeps (Figure 4 and Supplementary Table 5). One striking example is a haplotype of 25% frequency in YRI in the region containing the BAK1 and HLA-DPA1 genes (Supplementary Fig. 2). Further study of this and other haplotypes that have putatively undergone selection may point to key functional changes in the MHC that have influenced human disease past and present.
Figure 4.
The genetic distance over which the long-range haplotype associated with each allele for each SNP on chromosome 6 extends (before decaying to an EHH22 of 0.8) in each of the four populations. (See Methods for details.) The blue dot represents the average extent of long-range haplotypes for SNP alleles in 20 different frequency bins (0%-5% 5%-10%, etc...), with the 95% confidence interval represented by a black line. HLA alleles above the 95% confidence interval are presented by red diamonds.
We set out to create a dense haplotype map across the extended MHC in four population samples. This resource will facilitate the selection of informative tag SNPs to capture HLA and non-HLA variation, enabling a cost-effective means for conducting association studies in large patient samples, and thus provide a complementary approach to classical HLA typing. We anticipate that future integration with the efforts from the International HapMap Project, International Histocompatibility Working Group, and the MHC Haplotype Project combined with targeted functional studies will help identify the causal alleles that predispose to immune-mediated diseases and those that have been under selection23,24.
Methods
DNA samples
Our study includes 90 individuals (30 parent-offspring trios) of the Yoruba people from Ibadan, Nigeria (YRI); 182 Utah residents (29 extended families -average family size of 6.2 - containing 45 unrelated parent-offspring trios) with European ancestry from the Centre d’Etude du Polymorphisme Humain (CEPH) collection (CEU); 45 unrelated Han Chinese from Beijing, China (CHB); and 44 unrelated Japanese from Tokyo, Japan (JPT). These samples correspond to the 269 DNA samples used in Phase I of the International HapMap Project, plus an additional set of 92 CEU samples. Most of this expanded set of CEU samples were also included in our previous studies of the MHC2,3. To test tag transferability, we studied 330 samples from a celiac disease study conducted in The Netherlands13 and 996 samples from a UK systemic lupus erythematosus study14. The study was approved by the Medical Ethics Committee of the University Medical Center Utrecht and by the London multi-centre research ethics committee (MREC), respectively, and written informed consent was obtained from all the participants.
Typing of SNPs and insertion/deletion polymorphisms
SNPs and DIPs were identified from the MHC Haplotype Project, dbSNP, and dbMHC databases and selected based on their genomic position. SNPs were typed on the Illumina GoldenGate platform at the Broad Institute of MIT and Harvard, at Illumina, and at the Wellcome Trust Sanger Institute or by using using TaqMan Allelic Discrimination Assay at Duke University. Insertion/deletion polymorphisms were typed by TaqMan technology at Duke University. All of the SNP, DIP and HLA typing was completed by June 2005 and preceded the release of Phase II data from the International HapMap Project. The entire list of 7,543 non-redundant variants and their respective genotyping assays are available online (see below). The variants were located in the 7.5 Mb region delimited by rs498548 (position chr6:26000508) and rs2772390 (position chr6:33483033). All coordinates are given relative to NCBI build 34 of the human genome assembly. Raw genotype data collected at the various genotyping centers were collated based on map position. A total of 6,338 variants yielded reliable genotyping assays. Assays considered to be reliable yielded at least 90% total genotypes, fewer than two Mendel errors, and were in Hardy-Weinberg equilibrium (P > 0.001). Details of how haplotypes were estimated from genotype data can be found in Supplementary Note online.
HLA typing
HLA typing was carried out at the Laboratory of Genomic Diversity (NCI-FCRDC) using PCR-SSOP (sequence specific oligonucleotide probe) based protocols recommended by the 13th International Histocompatibility Workshop (http://www.ihwg.org/components/ssopr.htm). For class I HLA (A, B, and C) typing the gene fragment containing exon 2, intron 2, and exon 3 was selectively amplified using locus-specific primers. For the class II HLA (DQA1, DQB1 and DRB1) typing only exon 2 was examined. Genotype ambiguities were resolved by direct sequencing of the whole PCR fragment.
Statistical analysis of LD
To measure LD between biallelic markers we used the r2 measure of association26. To measure the strength of LD between a biallelic SNP, i, and the multi-allelic HLA locus, j, with N alleles, we use relative information, defined as
where
is the frequency of the kth HLA allele on the ‘x’ allele at the SNP (the lack of a superscript indicates unconditional frequencies) and is the frequency of the ‘x’ allele in the sample. To test for significant association between alleles at two HLA loci, we calculate a χ2 test statistic and obtain a P-value by permutation.
Selection of tag SNPs
We used the Tagger method11 to derive SNP-based tests to capture all observed HLA alleles in the four population samples. For each HLA allele, we first evaluate the maximum r2 for single-marker tests (based on a single tag). If the maximum r2 < 1.0, we proceed to evaluate multimarker tests based on multiple SNPs surrounding the HLA allele (up to 500 kb distance), and keep the haplotype test with the highest r2 to the HLA allele (Supplementary Table 3).
Empirical validation
We selected tag SNPs to capture the DQA1*0501 and DQA*0201 alleles (DQ2.5), the DQA1*0201 and DQB1*0202 alleles (DQ2.2), DRB1*1501 and DRB1*0301 in the CEU reference panel. We genotyped these tag SNPs and also performed classical HLA typing for these HLA alleles in the respective disease samples (celiac and SLE), allowing us to evaluate empirically how well these tags can predict the actual allelic state of these HLA genes in the patients. We report the sensitivity and the specificity of these SNP-based tests, as well as the empirical r2 between the test and the allele (Table 2).
Analysis of recombination and haplotype structure
Recombination rates and the location of recombination hotspots with strong statistical support were estimation from patterns of genetic variation using previously described methods27-29. Analyses were carried out separately on each analysis panel (YRI, CEU, CHB+JPT) and results were combined to provide a single genetic map for the region (Fig. 1). In addition, we identified (for each panel) all non-redundant haplotypes with a frequency of ≥10% and consisting of at least 10 SNPs, which are likely to represent the non-recombinant descendants from a single ancestor.
Estimation of evolutionary trees from haplotype data
We use a simple, heuristic approach to estimate the genealogical history at a given point, x, along the chromosome from a set of phased haplotypes (with missing data imputed). The algorithm is initialized by setting the age of each haplotype or lineage, ti, to zero. At each step of the algorithm we identify (and remove) all singleton mutations, recording the number that occur unambiguously (see below for a definition of unambiguous) on each lineage as si. We then calculate a statistic for each pair of haplotypes
where is the number of SNPs 5′ of position x until the first point at which haplotypes i and j differ ( is the equivalent number for SNPs 3′ of position x). We identify the pairs of haplotypes with the largest statistic, and select among those the pair of haplotypes with the fewest mutations (i.e. the smallest value of si + sj). This pair of haplotypes is coalesced, generating a new lineage, k, by generating ‘recombination’ events at the end points of identity in both haplotypes at both ends. The relative time at which the coalescent takes place is estimated from
where c is a constant (we use an arbitrary value of c = 100). Recombinant lineages that do not include position x are discarded and the process is repeated until a single lineage remains. The algorithm is repeated until a single lineage remains.
The recombination process results in the presence of non-ancestral material in lineages. When calculating identity, this is treated as ‘not identical’. Unambiguous association of a singleton mutation with a lineage is only allowed if all copies of the mutation to be removed have been removed through coalescence (i.e. not through the discarding of recombinants). Due to the heuristic nature of the algorithm, the estimated tree should only be taken as an approximation, but one that performs well in capturing recent haplotype history.
Search analysis for evidence of historical selection
We used four implementations of the Long Range Haplotype (LRH) test to examine evidence for recent positive selection in the HLA22. Long-range association is measured by extended haplotype homozygosity (EHH). For a population of individuals sharing allele t, EHH at a distance x from the locus is defined as the probability that two randomly chosen chromosomes carrying the allele of interest are identical by descent (as assayed by homozygosity at all SNPs)30 for the entire interval from the locus to the point x. The first two implementations were the traditional approach previously described 22 where the allele of interest was either a single SNP or a haplotype (between 3 and 10 SNPs). The third implementation was the integrated EHH method recently described31. These first three implementations use the other haplotypes present at the locus to control for local recombination rate, this approach might obscure evidence for recurrent selection at a locus. We therefore used a fourth implementation for each SNP in our data, which might be better suited for detecting recurrent sweeps. We measured the genetic distance the haplotype extends before decaying to an EHH of 0.827 (presented in Figure 4).
We visualize the decay of the extended ancestral chromosome (haplotype) on which the allele arose22 using the program Bifurcator32. The root of each diagram is an allele, identified by an open square. The diagram is bi-directional, portraying both centromere-proximal and centromere-distal LD. Moving in one direction, each marker is an opportunity for a node; the diagram either divides or not based on whether both or only one allele for each adjacent marker is present. Thus, the breakdown of LD away from the allele of interest is portrayed at progressively longer distances. The thickness of the lines corresponds to the number of samples with the indicated long-distance haplotype.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Drs. Jorge Oksenberg, Phil De Jager and Neil Walker for helpful discussions and their critical reading of the manuscript. The authors are also grateful to Ben Fry for technical assistance with the selection analysis. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The Wellcome Trust supported the work of M.M., P.W., M.D., J.M., S.B., J.T., J.A.T. and P.D. The Juvenile Diabetes Research Foundation supported J.A.T. The International MS Genetics Consortium supported the work of D.H., S.G., M.P.V., and JDR. This work was also supported by grants from the NIDDK and the NIAID (Autoimmunity Prevention Center Grant U19 AI050864) to JDR.
Footnotes
All data will be available at the following sites:
http://www.broad.mit.edu/mpg/idrg/projects/hla/
COMPETING FINANCIAL INTEREST STATEMENT
The authors declare that they have no competing financial interests.
References
- 1.Dupont B, Svejgaard A. HLA and disease. Transplant Proc. 1977;9:1271–4. [PubMed] [Google Scholar]
- 2.Miretti MM, et al. A high-resolution linkage-disequilibrium map of the human major histocompatibility complex and first generation of tag single-nucleotide polymorphisms. Am J Hum Genet. 2005;76:634–46. doi: 10.1086/429393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walsh EC, et al. An integrated haplotype map of the human major histocompatibility complex. Am J Hum Genet. 2003;73:580–90. doi: 10.1086/378101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allcock RJ, et al. The MHC haplotype project: a resource for HLA-linked association studies. Tissue Antigens. 2002;59:520–1. doi: 10.1034/j.1399-0039.2002.590609.x. [DOI] [PubMed] [Google Scholar]
- 5.Horton R, et al. Gene map of the extended human MHC. Nat Rev Genet. 2004;5:889–99. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 6.Stewart CA, et al. Complete MHC haplotype sequencing for common disease gene mapping. Genome Res. 2004;14:1176–87. doi: 10.1101/gr.2188104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malkki M, Single R, Carrington M, Thomson G, Petersdorf E. MHC microsatellite diversity and linkage disequilibrium among common HLA-A, HLA-B, DRB1 haplotypes: implications for unrelated donor hematopoietic transplantation and disease association studies. Tissue Antigens. 2005;66:114–24. doi: 10.1111/j.1399-0039.2005.00453.x. [DOI] [PubMed] [Google Scholar]
- 8.Stenzel A, et al. Patterns of linkage disequilibrium in the MHC region on human chromosome 6p. Hum Genet. 2004;114:377–85. doi: 10.1007/s00439-003-1075-5. [DOI] [PubMed] [Google Scholar]
- 9.Limm TM, Ashdown ML, Naughton MJ, McGinnis MD, Simons MJ. HLA-DQA1 allele and suballele typing using noncoding sequence polymorphisms. Application to 4AOHW cell panel typing. Hum Immunol. 1993;38:57–68. doi: 10.1016/0198-8859(93)90520-b. [DOI] [PubMed] [Google Scholar]
- 10.Simons MJ, et al. Strategy for definition of DR/DQ haplotypes in the 4AOHW cell panel using noncoding sequence polymorphisms. Hum Immunol. 1993;38:69–74. doi: 10.1016/0198-8859(93)90521-2. [DOI] [PubMed] [Google Scholar]
- 11.de Bakker PIW, et al. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–23. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez-Neira A, et al. The portability of tagSNPs across populations: a worldwide survey. Genome Res. 2006;16:323–30. doi: 10.1101/gr.4138406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monsuur AJ, et al. Myosin IXB variant increases the risk of celiac disease and points toward a primary intestinal barrier defect. Nat Genet. 2005;37:1341–4. doi: 10.1038/ng1680. [DOI] [PubMed] [Google Scholar]
- 14.Chadha S, et al. Haplotype analysis of tumour necrosis factor receptor genes in 1p36: no evidence for association with systemic lupus erythematosus. Eur J Hum Genet. 2006;14:69–78. doi: 10.1038/sj.ejhg.5201527. [DOI] [PubMed] [Google Scholar]
- 15.Vader W, et al. The HLA-DQ2 gene dose effect in celiac disease is directly related to the magnitude and breadth of gluten-specific T cell responses. Proc Natl Acad Sci U S A. 2003;100:12390–5. doi: 10.1073/pnas.2135229100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham RR, et al. Visualizing human leukocyte antigen class II risk haplotypes in human systemic lupus erythematosus. Am J Hum Genet. 2002;71:543–53. doi: 10.1086/342290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marsh SG. Nomenclature for factors of the HLA system, update June 2005. Tissue Antigens. 2005;66:338–40. doi: 10.1111/j.1399-0039.2005.00479.x. [DOI] [PubMed] [Google Scholar]
- 18.Klein J. Origin of major histocompatibility complex polymorphism: the trans-species hypothesis. Hum Immunol. 1987;19:155–62. doi: 10.1016/0198-8859(87)90066-8. [DOI] [PubMed] [Google Scholar]
- 19.Raymond CK, et al. Ancient haplotypes of the HLA Class II region. Genome Res. 2005;15:1250–7. doi: 10.1101/gr.3554305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Traherne JA, et al. Genetic Analysis of Completely Sequenced Disease-Associated MHC Haplotypes Identifies Shuffling of Segments in Recent Human History. PLoS Genet. 2006;2:e9. doi: 10.1371/journal.pgen.0020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Froeschke G, Sommer S. MHC class II DRB variability and parasite load in the striped mouse (Rhabdomys pumilio) in the Southern Kalahari. Mol Biol Evol. 2005;22:1254–9. doi: 10.1093/molbev/msi112. [DOI] [PubMed] [Google Scholar]
- 22.Sabeti PC, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–7. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 23.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 24.Wang WY, Barratt BJ, Clayton DG, Todd JA. Genome-wide association studies: theoretical and practical concerns. Nat Rev Genet. 2005;6:109–18. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- 25.Petersdorf EW, Malkki M. Human leukocyte antigen matching in unrelated donor hematopoietic cell transplantation. Semin Hematol. 2005;42:76–84. doi: 10.1053/j.seminhematol.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 26.Hill WG RA. Linkage disequilibrium in finite populations. Theor.Appl.Genet. 1968;38:226–231. doi: 10.1007/BF01245622. [DOI] [PubMed] [Google Scholar]
- 27.The International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McVean GA, et al. The fine-scale structure of recombination rate variation in the human genome. Science. 2004;304:581–4. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- 29.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–4. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 30.Nei M, editor. Molecular evolutionary genetics. Columbia University Press; New York: 1987. [Google Scholar]
- 31.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fry B. Thesis. Massachusetts Institute of Technology; 2005. Computational Information Design. [Google Scholar]
- 33.Kong A, et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–7. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.