Abstract
Peripheral artery disease (PAD) is a leading cause of cardiovascular morbidity and mortality1; however, the extent to which genetic factors increase risk for PAD is largely unknown. Using electronic health record data, we performed a genome-wide association study in the Million Veteran Program testing ~32 million DNA sequence variants with PAD (31,307 cases and 211,753 controls) across veterans of European, African, and Hispanic ancestry. The results were replicated in an independent sample of 5,117 PAD cases and 389,291 controls from UK Biobank. We identified 19 PAD loci, 18 of which have not been previously reported. 11 of the 19 loci were associated with disease in three vascular beds (coronary, cerebral, peripheral), including LDLR, LPL, and LPA, suggesting that therapeutic modulation of LDL cholesterol, the LPL pathway or circulating lipoprotein(a) may be efficacious for multiple atherosclerotic disease phenotypes. Conversely, 4 of the variants appeared to be specific for PAD, including F5 p.R506Q, highlighting the pathogenic role of thrombosis in the peripheral vascular bed and providing genetic support for Factor Xa inhibition as a therapeutic strategy for PAD. Our results highlight mechanistic similarities and differences among coronary, cerebral, and peripheral atherosclerosis and provide therapeutic insights.
Peripheral artery disease (PAD) is a complex disease impacted by both lifestyle and inheritance2. Despite its high prevalence, only a few studies have evaluated PAD genetics, with published genome-wide association studies (GWAS) having revealed only 3 loci reaching genome-wide significance3,4. Furthermore, it is uncertain if the genetic mechanisms underlying atherosclerotic disease of the peripheral arteries (PAD) and the coronary and cerebral arteries are shared or distinct.
Large-scale biobanks combining genetic data with electronic health record (EHR)-derived phenotypes are under development throughout the world5,6. The Million Veteran Program (MVP) was established in 2011 to study how genes affect health in the Veterans Affairs (VA) Healthcare System. Approximately 10% of individuals greater than the age of 55 seeking care in the VA Healthcare System have PAD, making MVP an ideal cohort for performing a large-scale PAD genetic analysis. Leveraging the MVP resource, we sought to: 1) perform a genetic discovery analysis for PAD; 2) explore the spectrum of phenotypic consequences associated with PAD risk variants; and 3) identify genetic signals that differentiate PAD from vascular disease in other arterial beds.
We designed a two-phased GWAS (Fig. 1). Initial discovery was performed in MVP, testing for association separately among individuals of European (whites), African (blacks), and Hispanic ancestry. The results were then meta-analyzed across ancestral groups. For variants with suggestive associations (P<10−6) with PAD, we sought replication in UK Biobank. We then combined statistical evidence across MVP and UK Biobank and set a significance threshold of P < 5 ×10−8 (genome-wide significance).
Figure 1. Discovery study design for the peripheral artery disease genome-wide association analysis.
Electronic health record based phenotyping identified 31,307 PAD cases of varying severity, as depicted in the upper row of boxes, in the Million Veteran Program. The association of DNA sequence variants with PAD was tested separately in 3 mutually exclusive ancestry groups and the results combined using an inverse-variance weighted fixed effects meta-analysis in the discovery phase. Variants with suggestive association (two-sided logistic regression P < 10−6) were then brought forward for independent replication in the UK Biobank.
Abbreviations: GWAS, genome-wide association study; PAD, Peripheral Artery Disease
The MVP discovery analysis was comprised of 31,307 individuals (24,009 white, 5,373 black, 1,925 Hispanic) with PAD and 211,753 disease-free controls; their baseline characteristics are presented in Supplementary Table 1. Participants with PAD were more likely to be older, male, prescribed statin therapy, have a history of smoking, and affected with type 2 diabetes (T2D). To validate our PAD phenotype, the minimum ankle-brachial index (mABI) was extracted for 17,861 individuals with ABI measurements available in MVP. As expected, we observed a median mABI of less than 0.9 for PAD cases and approximately 1 for PAD controls across all three ethnic groups (Supplementary Table 2, Extended Data Fig. 1). We further validated our MVP PAD phenotype with manual chart review and observed a specificity of 88% (95% CI = 75.7–94.5%) and sensitivity of 100% (95% CI = 89.8–100%), commensurate with that published in the literature7.
Through genotype imputation, we obtained 20.3 million, 32.4 million, and 31.2 million DNA sequence variants for analysis in white, black, and Hispanic participants, respectively (Supplementary Table 1). Following trans-ethnic meta-analysis in the discovery phase, a total of 554 variants at 25 loci met a genome-wide significance threshold (Extended Data Fig. 2). We replicated all 3 previously described genome-wide PAD loci with at least nominal (P < 0.05) significance (Supplementary Table 3). A total of 1,276 variants demonstrated association P<10−6 in the MVP discovery analysis. Of those, 552 were also available for independent testing in UK Biobank (5,117 PAD cases, 389,291 controls) and were taken forward for replication. Following replication, 19 loci exceeded genome-wide significance (P < 5×10−8, Table 1, Supplementary Table 4). Of the 19 PAD loci, 15 were directionally consistent across whites, blacks, and Hispanics in MVP, 8 demonstrated at least nominal significance in blacks and 3 in Hispanics (Supplementary Table 5); 18 of the loci have not been previously reported (Extended Data Fig. 3).
Table 1 -.
PAD risk loci discovered in the MVP biobank and replicated in the UK Biobank.
| Chr:Pos | rsid | EA | NEA | EAF | *Overall OR | *Overall 95% CI | *Overall P | Annotation | Gene/Locus** |
|---|---|---|---|---|---|---|---|---|---|
| 1:109817192 | rs7528419 | A | G | 0.772 | 1.07 | 1.05–1.09 | 2.54E-11 | 3’ UTR variant | CELSR2/SORT1 |
| 1:169519049 | rs6025 | T | C | 0.026 | 1.2 | 1.14–1.26 | 1.63E-12 | Missense variant (Factor V Leiden) | F5 |
| 6:160985526 | rs118039278 | A | G | 0.068 | 1.26 | 1.22–1.30 | 1.57E-43 | Intron variant | LPA |
| 6:31065071 | rs3130968 | T | C | 0.144 | 1.07 | 1.05–1.10 | 3.16E-10 | Regulatory region variant | (HLA-B) |
| 7:19049388 | rs2107595 | A | G | 0.187 | 1.08 | 1.05–1.10 | 2.49E-11 | Regulatory region variant | (HDAC9) |
| 7:22786532 | rs4722172 | G | A | 0.202 | 1.08 | 1.05–1.10 | 3.65E-11 | Intergenic variant | (IL6) |
| 8:19819217 | rs322 | A | C | 0.706 | 1.06 | 1.04–1.07 | 2.53E-09 | Intron variant | LPL |
| 9:136149229 | rs505922 | C | T | 0.334 | 1.06 | 1.04–1.07 | 7.10E-11 | Intron variant | ABO |
| 9:22103183 | rs1537372 | T | G | 0.421 | 1.12 | 1.10–1.14 | 4.32E-39 | Intron variant | CDKN2B-AS1/9p21 |
| 10:114758349 | rs7903146 | T | C | 0.293 | 1.06 | 1.04–1.08 | 3.76E-11 | Intron variant | TCF7L2 |
| 11:102710471 | rs566125 | T | C | 0.127 | 1.08 | 1.05–1.11 | 4.37E-09 | Intron variant | MMP3 |
| 11:46342834 | rs7476 | C | A | 0.364 | 1.06 | 1.04–1.08 | 8.33E-10 | 3’ UTR variant | CREB3L1 |
| 12:112871372 | rs11066301 | G | A | 0.413 | 1.06 | 1.04–1.08 | 2.96E-11 | Intron variant | PTPN11 |
| 12:79951566 | rs4842266 | G | A | 0.388 | 1.06 | 1.04–1.08 | 1.01E-09 | Upstream gene variant | RP11–359M6.3 |
| 13:110828891 | rs1975514 | C | T | 0.357 | 1.05 | 1.04–1.07 | 8.32E-10 | Intron variant | COL4A1 |
| 14:70501364 | rs55784307 | A | C | 0.183 | 1.06 | 1.04–1.09 | 2.93E-08 | Downstream gene variant | SMOC1 |
| 15:78915864 | rs10851907 | A | G | 0.41 | 1.06 | 1.05–1.08 | 1.49E-13 | Upstream gene variant | CHRNA3 |
| 17:66089393 | rs62084752 | C | G | 0.216 | 1.07 | 1.05–1.09 | 1.58E-10 | Upstream gene variant | LOC732538 |
| 19:11191729 | rs138294113 | C | T | 0.879 | 1.09 | 1.06–1.11 | 1.20E-10 | Intergenic variant | (LDLR) |
Overall OR, 95% CI, and P (two-sided) represent logistic regression statistics following meta-analysis of MVP and UK Biobank (total N = 36,424 PAD cases and 601,044 controls)
Genes for variants that are outside the transcript boundary of a protein-coding gene are shown with nearest candidate gene in parentheses [eg, (LDLR)].
Abbreviations: Chr, Chromosome; Pos, Position; rsid, RefSNP identification number; EA, Effect Allele; NEA, Non Effect Allele; EAF, Effect Allele Frequency; OR, Odds Ratio; CI, Confidence Interval
The LPA variant rs118039278 was the top association result (6.4% frequency for the A allele; OR =1.25; 95%CI: 1.22–1.30; P = 1.57×10−43). Of the 6 signals from MVP that did not replicate in the UK Biobank, 2 were rare variants that were not available in UK Biobank following quality control (European MAF < 0.005), and the remaining four did not meet the pre-specified P < 0.05 for independent replication (Supplementary Table 6). All 3 previously reported suggestive (5.0×10−8 < P < 0.05) PAD associations at the SH2B3/PTPN118, HDAC94, and CHRNA39 loci were observed at genome-wide significance.
We next sought to determine whether DNA sequence variants were associated with PAD severity as determined by mABI. We performed a GWAS of mABI as a continuous trait for 13,382 European, 3,284 African, and 998 Hispanic ancestry individuals in MVP, restricting to those with an ABI < 1.4 as previously described3. Baseline characteristics for these individuals are depicted in Supplementary Table 7. Following trans-ethnic meta-analysis, only the known 9p21-ABI association3 passed the genome-wide significance threshold [rs1333045, 46.8% frequency for the T allele; β = 0.064; 95%CI: 0.042–0.086; P = 8.3×10−9]. However, we observed that 6 of the 19 PAD risk variants identified in our PAD case/control analysis were associated with reduced mABI at nominal significance (P < 0.05, Supplementary Table 8). Notably, the mABI GWAS lead 9p21 variant (rs1333045) was different than the lead variant identified in the PAD case-control analysis at this locus (rs1537372).
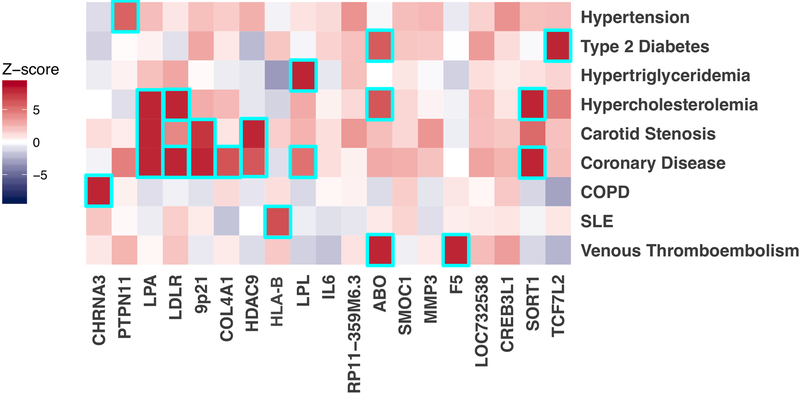
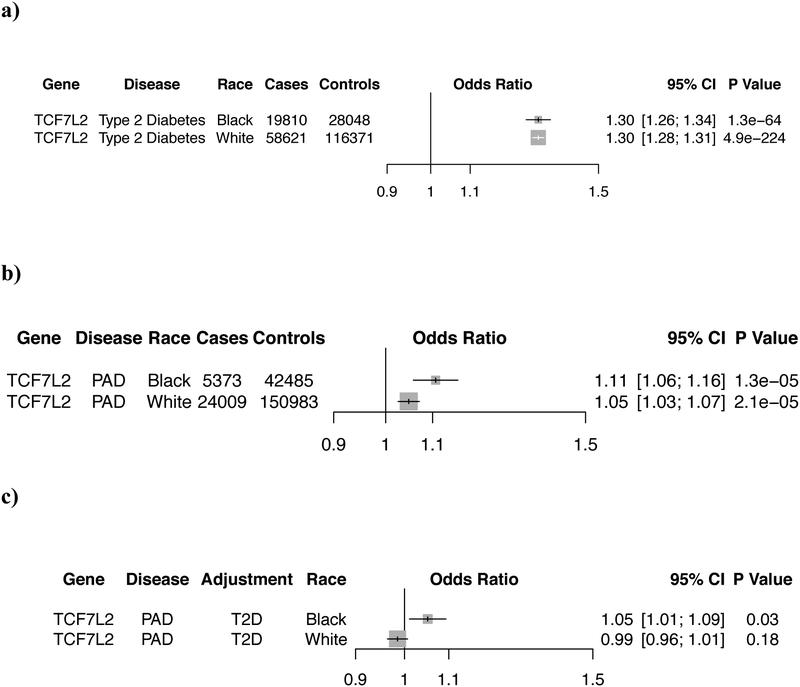
Understanding the full spectrum of phenotypic consequences of a given DNA sequence variant can help identify the mechanism by which a variant or gene leads to disease. Termed a phenome-wide association study (PheWAS), this approach examines the association of a risk variant across a range of phenotypes10,11. Using a median of 65 distinct ICD-9/10 EHR-derived diagnosis codes per participant, we tested each of the 19 PAD lead risk variants across 1,101 disease phenotypes. We found that several of the newly identified DNA sequence variants correlated with a range of known risk factors for PAD (Fig. 2, Supplementary Table 9). For example, rs7903146 within TCF7L2 is one of the strongest known genetic predictors of T2D12 and associated with T2D in our PheWAS. The PAD association for rs7903146 was significantly reduced when controlling for T2D in the regression model, suggesting this variant confers PAD risk through its effect on T2D (Extended Data Fig. 4). The Factor V Leiden variant (F5 p.R506Q) demonstrated a known association with venous thromboembolism13. We found four PheWAS associations with hypercholesterolemia and one with hypertriglyceridemia. These loci have previously associated with either LDL cholesterol (LDLR, ABO, SORT1, LPA) or triglycerides (LPL)14, known causal paths to atherosclerosis. rs10851907 in CHRNA3 (encoding cholinergic receptor nicotinic alpha-3) demonstrated an association with chronic obstructive pulmonary disease. This DNA sequence variant is strongly correlated (R2 = 0.73) with variants previously shown to predict nicotine dependence9 and appears to drive PAD risk entirely through its effect in smokers (Extended Data Fig. 5). rs3130968 near the HLA-B gene was associated with a number of autoimmune diseases including Celiac disease, Graves’ disease, Systemic Lupus Erythematosus, and type 1 diabetes15. In total, we identified 158 statistically significant (P < 5.0×10−8) PheWAS associations across the 19 genetic variants implicating many known PAD risk factors based on the traits they relate to - including lipids, type 2 diabetes, smoking, thrombosis, and hypertension16 (Extended Data Fig. 6).
Figure 2. Representative heatmap of phenome-wide association results and biologic pathways underlying genetic loci associated with peripheral artery disease.
Logistic regression Z-scores (aligned to the PAD risk allele) from the MVP PheWAS analysis (N = 176,913) or publically available PheWAS results from PhenoScanner 2.0 (variable N, see Supplementary Table 23) are shown for the associations between the 19 PAD risk loci and representative disease traits. A positive Z-score (red) indicates a positive association between the PAD risk allele and the disease, whereas a negative Z-score (blue) indicates an inverse association. Boxes are outlined in cyan if the variant is associated with the indicated disease at genome-wide significance (logistic regression two-sided P < 5.0 ×10−8).
Abbreviations: COPD, chronic obstructive pulmonary disease; SLE, Systemic Lupus Erythematosus
We supplemented our MVP PheWAS using data from PhenoScanner V217, an online resource of association statistics from previously conducted GWAS and UK Biobank. In total, we identified 443 additional PheWAS associations from the PhenoScanner database at P < 5 ×10−8 (Supplementary Table 10). We subsequently prioritized likely candidate causal PAD risk genes by aggregating evidence from i) prior genetic, clinical, or functional studies, ii) our PheWAS results, iii) cis-eQTLs from the Genotype-Tissue Expression Project (GTEx) V7 dataset18, iv) recently published pQTL data derived from the human plasma of 3,301 participants of the INTERVAL study19,20, and v) results from a transcriptome-wide association study21 (TWAS) using RNA-seq data from post-mortem tibial artery tissue (388 individuals) and MVP European PAD summary statistics. This analysis revealed several candidate causal genes including F5, LPA, SORT1, LPL, and LDLR (Supplementary Tables 11,12).
We next sought to better understand how DNA sequence variants might differ in their contribution to vascular disease risk in the peripheral, coronary, and cerebral arterial territories. Analysis of shared heritability provides a mechanism to better understand the relationship of common variant risk across phenotypes22,23. Using linkage disequilibrium score regression23, we examined the genetic correlation between PAD and both coronary artery disease (CAD) and large artery stroke (LAS). We used summary statistics from the European MVP PAD analysis, along with summary data of 60,801 coronary disease cases and 123,504 disease-free controls from the CARDIoGRAMplusC4D consortium24, and 6,688 LAS cases and 454,450 controls from the 2018 MEGASTROKE analysis25. We noted a stronger positive correlation between PAD and LAS (rg = 0.88, P = 5.5×10−6) than for PAD and CAD (rg = 0.62, P = 1.57×10−43). Based on these findings, we sought to further explore the differential effects of individual genetic variants on PAD, LAS, and CAD.
For the 19 lead PAD risk variants identified in our GWAS analysis, we first tested their effects on CAD and LAS in white MVP participants and then combined the results with summary statistics from the CARDIoGRAMplusC4D or MEGASTROKE studies, respectively. We observed that 14 PAD risk variants demonstrated at least nominal association (P < 0.05) with CAD, and 12 with LAS (Supplementary Tables 13–16). In a sensitivity analysis, the PAD effect estimates at the SORT1, LPA, 9p21, and LDLR loci were attenuated, suggesting that some of the PAD risk may be driven by comorbidity or shared causal pathways when accounting for the concomitant CAD and LAS diagnoses (Supplementary Tables 17–19). Interestingly the COL4A1 locus, previously associated with CAD24 and small vessel disease of the brain26, was found to be associated with PAD and CAD but not LAS in our analysis. Data from the MEGASTROKE study demonstrate evidence of association with small artery stroke (P = 1.4×10−4) for this variant, suggesting it may be acting differently in the cerebral bed.
Common mechanisms emerged for the 11 PAD risk variants demonstrating significant association in all three (coronary, cerebral, peripheral) vascular beds including lipids (LDLR, LPA, LPL, SORT1), hypertension (PTPN11), and diabetes (TCF7L2). Conversely, variants in the RP11-359M6.3, HLA-B, CHRNA3, and F5 loci were uniquely associated with PAD, implying that smoking and thrombosis may play a greater role in PAD than disease in other arterial territories (Extended Data Fig. 7–8).
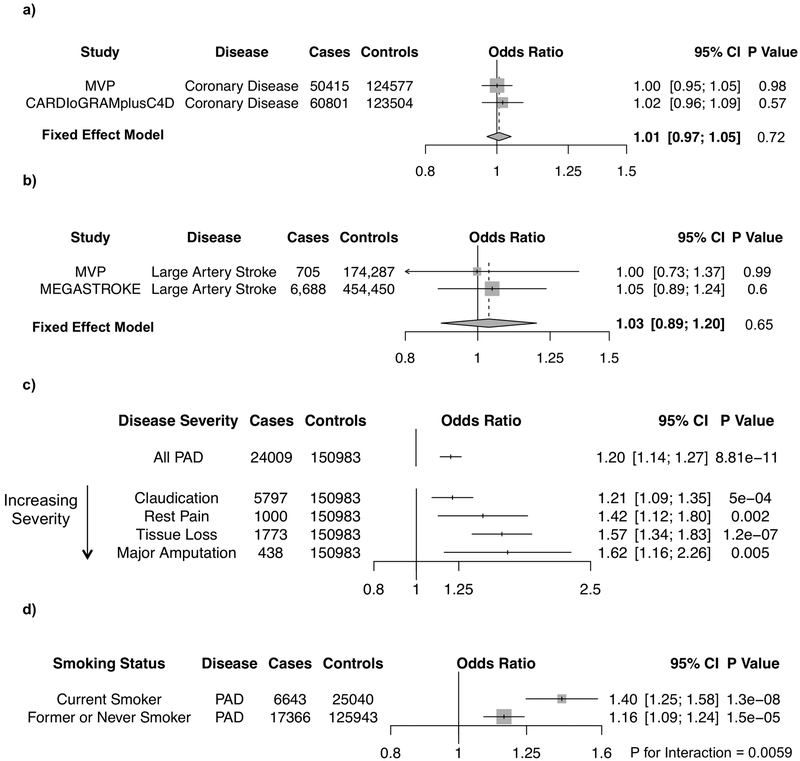
The novel PAD risk variant Factor V Leiden (F5 p.R506Q) is the most common cause of inherited thrombophilia27, as the variant’s protein-altering consequence results in a resistance to proteolysis by activated protein C28. In a combined analysis of 111,216 coronary disease cases and 248,081 controls from MVP (9,388 Factor V Leiden carriers) and CARDIoGRAMplusC4D, we observed no evidence of an association between F5 p.R506Q and CAD (OR =1.01; 95%CI: 0.97–1.05; P =0.72, Fig. 3a). Similarly, for 7,393 LAS cases and 628,737 controls from MVP and MEGASTROKE, we observed no evidence of an association between F5 p.R506Q and LAS (OR =1.03; 95%CI: 0.89–1.20; P =0.65, Fig. 3b). In contrast, F5 p.R506Q was associated with a 20% increased risk of PAD in individuals of European ancestry in MVP (OR =1.20; 95%CI: 1.14–1.27; P =8.81×10−11, Fig. 3c).
Figure 3. Factor V Leiden mutation and vascular disease.
(a–d) The association of the thrombophilic Factor V Leiden variant, F5 p.R506Q, with different types of vascular disease were analyzed, as depicted in forest plots. Associations are shown with CAD (a) and LAS (b) using MVP and GWAS meta-analysis data (either CARDIoGRAM plusC4D or MEGASTROKE, respectively) that was combined using an fixed-effects, inverse-variance weighted meta-analysis. Associations with all PAD cases, as well as PAD cases of increasing severity (c), and PAD cases stratified by smoking status (d) among European ancestry MVP participants are shown. Two-sided logistic regression P values are displayed. Gray boxes reflect the inverse-variance weight for each study or subgroup.
Abbreviations: CI, Confidence Interval; CAD, Coronary Artery Disease; LAS, Large Artery Stroke; PAD, Peripheral Artery Disease; MVP, Million Veteran Program; GWAS, Genome-wide Association Study
To better understand Factor V Leiden’s relationship with PAD, we tested its association with increasingly severe disease manifestations, including claudication, rest pain, tissue loss, and major amputation. In total, we identified 5,797 individuals with intermittent claudication, 1,000 with rest pain, 1,773 with evidence of tissue loss, and 438 who had undergone a major amputation among white MVP participants (Supplementary Table 2). We observed significant associations for the Factor V Leiden mutation with each subtype of PAD (Fig. 3c). Interestingly, the variant’s effect estimate increased as PAD severity increased, with carriers having a 62% increased risk of undergoing a PAD-related major amputation (OR =1.62; 95%CI: 1.16–2.26; P =0.005).
Recent evidence has linked tobacco use to an increased risk for thrombotic sequelae29,30. We hypothesized that there may be an interaction between smoking and F5 p.R506Q carrier status on PAD risk. We observed that the presence of F5 p.R506Q had greater effect on PAD among current smokers (OR =1.40; 95%CI: 1.25–1.58; P = 1.3×10−8) than among former or never smokers (OR =1.16; 95%CI: 1.09–1.24; P = 1.5×10−5) (Cochran Q interaction two-sided P =0.0059, Fig. 3d). These findings may be secondary to a synergistic effect of active tobacco consumption on the hypercoagulability induced by F5 p.R506Q.
Our study should be interpreted within the context of its limitations. First, our PAD phenotype is based on EHR data and may result in misclassification of case status. Such misclassification should, however, reduce statistical power for discovery and on average bias results toward the null. Second, the VA Healthcare System population is overwhelmingly male, and although over 20,000 women were included in our analysis, our ability to detect sex-specific genetic associations was limited. Third, our mABI values were extracted from the EHR using natural language processing techniques from unstructured data, and these values are subject to greater misclassification than those ascertained from a prospective cohort study. Lastly, while we maximized the number of participants in our PheWAS analysis, it may still have been underpowered to detect association with certain diseases.
These findings permit several conclusions. First, a multi-ethnic, VA Healthcare System-based biobank offers potential to aid genetic discovery for understudied atherosclerosis syndromes. Previously published genome-wide PAD efforts have been limited by small sample sizes4, and in our study we leverage the high prevalence of atherosclerotic disease within the VA Healthcare System31 to increase the number of PAD cases analyzed by 10-fold. The extensive VA EHR - including a median of 10.0 years of follow-up per participant, >21 million prevalent diagnosis codes, and 261,835 ABI measurements - enabled us to identify and validate PAD cases, more deeply phenotype patients with sequelae of severe PAD, and highlight causal mechanisms of PAD risk variants through PheWAS. Our findings provide genetic evidence that therapies targeting atherosclerotic risk factors are likely to mitigate the rising incidence of PAD32.
Second, our results highlight mechanistic symmetries and differences between coronary, cerebral, and peripheral vascular disease that provide therapeutic insights. We identified 11 genetic loci common to CAD, LAS, and PAD, including the low-density lipoprotein receptor (LDLR), lipoprotein lipase (LPL), and lipoprotein(a) (LPA). These data suggest that therapeutic modulation of LDL cholesterol, the LPL pathway, or circulating lipoprotein(a)33–36 may all be efficacious for atherosclerosis in multiple vascular beds, including PAD. Conversely, the identification of four genetic signals specific to PAD imply that certain therapies may produce a substantially greater therapeutic benefit in one vascular bed over another and rejuvenate hypotheses regarding the role of autoimmune disease in atherosclerosis37. Further genetic analysis with greater sample sizes may reveal additional therapeutic targets that uniquely benefit PAD patients.
Third, our findings lend human genetic support to targeting the coagulation cascade as a therapeutic strategy for PAD. In our study, carriers of the thrombophilic Factor V Leiden mutation demonstrated a significantly increased risk of severe PAD including rest pain, tissue loss, and major amputation. Recent results from the COMPASS trial are consistent with our genetic findings, having demonstrated that the addition of low-dose rivaroxaban to aspirin prevented major adverse limb events including major amputation38. Rivaroxaban selectively inhibits factor Xa and in the COMPASS trial was used at levels well below the antithrombotic dose suggesting that there may be something specific about direct factor Xa inhibition that prevents adverse limb outcomes. Studies like ours provide additional mechanistic support to this hypothesis, given the intimate relationship between factors Xa and V in the thrombotic cascade; factor Xa activates factor V, and factor Va is a prerequisite for factor Xa to convert prothrombin to thrombin suggesting a potentially important mechanism of limb atherogenesis.
In summary, we identified 18 novel genomic loci associated with PAD risk, explored the phenotypic consequences of PAD risk variants through PheWAS, and identified 4 risk variants that appear to drive vascular disease more specifically in the peripheral vasculature, including the Factor V Leiden variant. These results are demonstrative of how large biobanks that couple genetic variation with dense EHR data can be leveraged for biological insights that can inform clinical care.
Online Methods
Study Populations
We conducted genetic association analyses using DNA samples and phenotypic data from two cohorts: MVP and UK Biobank. In MVP, individuals aged 19 to over 100 years have been recruited from 63 VA Medical Centers across the United States. In our initial MVP analysis, we evaluated 31,307 individuals (24,009 white, 5,373 black, 1,925 Hispanic) with PAD, and 211,753 controls free of clinical evidence of disease. For variants with suggestive associations (P < 10−6), we sought replication of our findings in UK Biobank (Fig. 1, Extended Data Fig. 9). In UK Biobank, individuals aged 45 to 69 years old were recruited from across the United Kingdom for participation. In this study, we identified 5,117 PAD cases and 389,291 controls of European ancestry. MVP received ethical/study protocol approval by the VA Central Institutional Review Board, the analysis in UK Biobank was approved by a local Institutional Review Board at Partners Healthcare (protocol 2013P001840), and informed consent was obtained for all participants. Additional information regarding experimental design and participants are provided in the Life Sciences Reporting Summary.
Genetic Data and Quality Control
DNA extracted from whole blood was genotyped in MVP using a customized Affymetrix Axiom biobank array, the MVP 1.0 Genotyping Array. Veterans (U.S. military personnel) of three mutually exclusive ethnic groups were identified for analysis: 1) non-Hispanic whites (European ancestry), 2) non-Hispanic blacks (African ancestry), and 3) self-identified Hispanics. Prior to imputation, variants that were poorly called or that deviated from their expected allele frequency based on reference data from the 1000 Genomes Project39 were excluded. After pre-phasing using EAGLE v240, genotypes from the 1000 Genomes Project39 phase 3, version 5 reference panel were imputed into Million Veteran Program (MVP) participants via Minimac3 software41. Ethnicity-specific principal component analysis was performed using the EIGENSOFT v6 software42. Participants were then divided into three mutually exclusive ethnic groups based on self-identified race/ethnicity and admixture analysis using the ADMIXTURE v1.3 software43: 1) non-Hispanic whites (self-identified as “non-Hispanic,” “white,” and > 80% genetic European ancestry), 2) non-Hispanic blacks (self-identified as “non-Hispanic,” “black,” and > 50% genetic African ancestry), and 3) Hispanics (self-identified only). In total, 312,571 white, black, and Hispanic MVP participants passed our sample-level quality control.
In MVP, sample and variant quality control was performed as previously described44. In brief, duplicate samples, samples with more heterozygosity than expected, an excess (>2.5%) of missing genotype calls, or discordance between genetically inferred sex and phenotypic gender were excluded. In addition, one individual from each pair of related individuals (kinship > 0.0884 as measured by the KING45 2.0 software) were removed.
Following imputation, variant level quality control was performed using the EasyQC R package46 (www.R-project.org), and exclusion metrics included: ancestry specific Hardy-Weinberg equilibrium47 P <1×10−20, posterior call probability < 0.9, imputation quality < 0.3, minor allele frequency (MAF) < 0.0003, call rate < 97.5% for common variants (MAF > 1%), and call rate < 99% for rare variants (MAF < 1%). Variants were also excluded if they deviated > 10% from their expected allele frequency based on reference data from the 1000 Genomes Project39. Following variant level quality control, we obtained 20.3 million, 32.4 million, and 31.2 million DNA sequence variants for analysis in white, black, and Hispanic participants, respectively.
In UK Biobank, analysis was performed separately in white individuals after genotyping using either the UK BiLEVE or UK Biobank Axiom Arrays. Approximately 500,000 individuals were genotyped and subsequently imputed to the haplotype reference consortium (HRC). Details of these procedures are described elsewhere48. We performed genome-wide association testing for PAD in the UK Biobank using all variants in the HRC reference with MAF > 0.5% and imputation quality INFO > 0.3. To avoid potential population stratification, only European-ancestry samples were included in the analysis. This subset was selected based on self-reported white ethnicity that was subsequently confirmed using genetic principal components analysis. Outliers within the self-reported white samples in the first 6 principal components of ancestry were detected and subsequently removed using the R package aberrant49. In addition, individuals with sex chromosome aneuploidy (neither XX or XY), discordant self-reported and genetic sex, or excessive heterozygosity or missingness, as defined centrally by the UK Biobank were removed. Finally, one individual from each pair of second-degree or closer relatives (kinship > 0.0884) was removed, selectively retaining PAD cases when possible.
Peripheral Artery Disease Definitions
From the 312,571 multi-ethnic participants passing quality control in MVP, individuals were defined as having PAD based on possessing at least two of the ICD-9/10 codes/CPT codes outlined in Supplementary Table 20 in their EHR, or having 1 code and at least 2 visits to a vascular surgeon within a 14 month period50. Individuals were defined as controls if they had zero diagnosis/procedure codes suggesting a diagnosis of PAD (including those in Supplementary Table 21) and their EHR reflected 2 or more separate encounters in the VA Healthcare System in each of the two years prior to enrollment in MVP. Manual chart review was performed by two trained nurse chart abstractors with a vascular surgeon reviewing discordant cases; the results of chart review for 50 cases and 50 controls otherwise representative of the overall cohort were used for determining the sensitivity and specificity of the phenotyping algorithm. In UK Biobank, individuals were defined as having PAD based on at possessing at least one of the self-reported illness codes, OPCS procedure codes, or ICD codes in Supplementary Table 22 in their EHR. All other individuals were defined as controls. In both cohorts, individuals were not excluded from the PAD control group if they possessed diagnosis codes for either CAD or LAS.
Assignment of Smoking Status in MVP
Smoking status was derived from an algorithm that utilized diagnosis codes, medications, clinic identifier codes, and smoking-related health factors from the VA EHR to classify individuals as never, former, or current smokers from American Heart Association abstract A18809 (http://circ.ahajournals.org/content/134/Suppl_1/A18809).
Ankle-Brachial Index Extraction and GWAS Quality Control
A natural language processing algorithm was used to extract ABI data from the EHR in MVP. Resultant values were manually inspected for accuracy. In total, 261,835 ABI measurements across 17,861 individuals were available for analysis. We selected each individual’s minimum ABI (mABI) for association analysis to minimize confounding from treatment or revascularization.
We performed a GWAS of mABI in 13,382 European, 3,284 African, and 998 Hispanic ancestry MVP participants after restricting to those with value < 1.4 as previously described3. Sample and variant quality control was performed in identical manner to the MVP PAD case/control analysis, with the exception of excluding variants with a MAF < 0.01 given the smaller sample size. In total, we obtained 9.2 million, 15.6 million, and 10.8 million DNA sequence variants for analysis in white, black, and Hispanic participants, respectively.
PheWAS of PAD Risk Variants
Understanding the full spectrum of phenotypic consequences of a given DNA sequence variant may shed light on the mechanism by which a variant/gene leads to disease. For lead PAD risk variants identified in our primary analysis, we performed a PheWAS of 1,101 distinct diseases in MVP leveraging the full catalog of EHR ICD-9 diagnosis codes in 176,913 white veterans passing PheWAS quality control using the R package PheWAS51 and its associated disease definitions with the exception of coronary artery disease defined as previously described52. Diseases were required to have a prevalence of > 0.2% (~300 cases) to be included in the PheWAS analysis. Lead PAD risk DNA sequence variants were tested using logistic regression adjusting for age, sex, and five principal components under the assumption of additive effects.
We supplemented our MVP PheWAS with data from PhenoScanner V217, an online resource of association statistics from previously conducted GWAS and UK Biobank and used a genome-wide significant P value threshold (two-sided P < 5 ×10−8). PhenoScanner data sources are outlined in Supplementary Table 23.
eQTL/pQTL associations and PAD Transcriptome-wide Association Study
To identify loci that might influence gene expression, we used previously published cis-expression quantitative trait locus (eQTL) mapping data from the Genotype-Tissue Expression (GTEx) Consortium Project across 44 tissues18. We queried the 19 PAD risk variants identified in our study for overlap with genome-wide significant variant-gene pairs from the GTEx portal. Similarly, to identify loci that might influence protein concentrations in plasma, we used published protein quantitative trait locus (pQTL) data generated from an aptamer-based multiplex protein assay to quantify 3,622 plasma proteins in 3,301 healthy participants from the INTERVAL study19,20. We queried the 19 lead PAD risk variants identified in our study for overlap with genome-wide significant variant-protein pairs.
We then performed a TWAS using summary statistics from the European MVP PAD analysis and gene-expression reference panels of tibial artery from GTEx V7 in 388 independent samples as previously described21. In brief, for a given gene, variant-expression weights in the 1-mB cis locus were first computed with the BSLMM53, which: “models effects on expression as a mixture of normal distributions to account for the sparse expression architecture. Given weights w, lipid Z scores Z, and variant-correlation (LD) matrix D; the association between predicted expression and lipids (i.e., the TWAS statistic) was estimated as ZTWAS = w’Z/(w’Dw)1/2 (details in ref.21).” We computed TWAS statistics by using either the variants genotyped in each expression reference panel or imputed HapMap3 variants. To account for multiple hypotheses we applied a Bonferroni corrected two-sided P < 6.2×10−6 = [0.05/8089 genes].
Shared Heritability within PAD, CAD, and LAS
To better understand the how common genetic variation influences risk for atherosclerosis in multiple vascular beds, we used linkage disequilibrium score regression23 to calculate the genetic correlation between PAD-CAD and PAD-LAS. Summary statistics from the European MVP PAD GWAS, the CARDIoGRAMplusC4D CAD GWAS24 (predominantly European), and the trans-ancestral LAS MEGASTROKE GWAS meta-analysis (>2/3 European)25 were used for this analysis. Of note, we used the trans-ancestral meta-analysis statistics from MEGASTROKE because the sample size of the European-ancestry only analysis lacked sufficient power for estimation of genetic correlation.
PAD Associations Independent of CAD and LAS
We sought to better understand how DNA sequence variants might differ in contribution to risk for atherosclerosis in the peripheral, coronary, and cerebrovascular beds. For 19 lead PAD risk variants identified in our primary analysis, we first tested their effect on CAD and LAS in white MVP participants and combined results with summary statistics from the CARDIoGRAMplusC4D and MEGASTROKE consortium studies, respectively. We performed a sensitivity analysis for variants demonstrating at least a nominally significant association with either CAD and/or LAS, by re-testing their association with PAD after including CAD or LAS status as a covariate in the association model. We also re-tested their association including both CAD and LAS as covariates in a single model. Variants associated with PAD, CAD, and LAS individually, and that remained associated with PAD after adjustment for CAD/LAS suggest the presence of a common mechanism or pathway leading to the development of atherosclerosis in multiple arterial beds. Conversely, associations that are present uniquely with PAD suggest a mechanism specific to the peripheral vascular bed.
MVP Coronary Artery Disease Definition and Analysis
CAD was defined based on ICD-9/10 and CPT codes using the method described by Dewey and colleagues52. CAD cases were defined as individuals who, based on ICD-9, ICD-10, and CPT codes had an inpatient admission with a primary diagnosis of CAD, a combination of CAD associated inpatient or outpatient encounters on two or distinct days noted in the longitudinal VA EHR or fee-for-service data, or a coronary revascularization at the time of analysis (Supplementary Table 24). We identified 50,415 CAD cases and 124,577 controls available for analysis among 174,992 white MVP participants. Genotyped and imputed DNA sequence variants were tested for association with CAD through logistic regression adjusting for age, sex, and five principal components of ancestry. Results were then combined with publicly available summary data of 60,801 CAD case patients and 123,504 disease free controls in the CARDIoGRAMplusC4D consortium study24 using an inverse-variance weighted fixed effects method. For variants with a high amount of heterogeneity across the two studies (I2 statistic > 75%, e.g., rs4842266), results were combined using a random effects method.
MVP Large Artery Stroke Adjusted Analysis and Definition
LAS was defined based on the groupings proposed by Denny et al51 and the phecode 433.11 - occlusion of the cerebral arteries with cerebral infarction, which is defined as the occurrence of the any of following ICD-9-CM codes on 2 distinct dates: 433.01, 433.11, 433.21, 433.31, 433.81, 433.91. We identified 705 LAS cases and 174,287 controls available for analysis among 174,992 white MVP participants. Genotyped and imputed DNA sequence variants were tested for association with LAS through logistic regression adjusting for age, sex, and five principal components of ancestry. Results were then combined with publicly available summary data of the transancestral LAS MEGASTROKE meta-analysis25 of 6,688 LAS cases and 454,450 controls using an inverse-variance weighted fixed effects method.
Type 2 Diabetes Definition for TCF7L2 adjusted analysis
To better understand how the TCF7L2 locus affects PAD risk, rs7903146 was re-tested for association with PAD after adjusting for type 2 diabetes (T2D) status in the 174,992 white MVP participants. T2D was defined based on the groupings proposed by Denny et al51, which identified 78,431 MVP participants affected with T2D (58,621 white and 5,273 black). We first tested the association of rs7903146 in MVP with T2D through logistic regression adjusting for age, sex, and five principal components of ancestry separately in whites and blacks. We then re-tested for association with PAD through logistic regression adjusting for age, sex, T2D, and five principal components of ancestry. We report logistic regression two-sided P values.
Factor V Leiden Genotypes and Risk of Vascular Disease
One of the variants most strongly associated with PAD in the discovery analysis was the Factor V Leiden mutation, the most common cause of inherited thrombophilia27. The variant’s protein altering consequence (F5 p.R506Q) results in a resistance to proteolysis by activated protein C and a hypercoaguable state28. We sought to better understand Factor V Leiden’s relationship with atherosclerosis by testing its association with CAD, LAS, and increasingly severe presentations of PAD. Individuals were defined as having claudication, rest pain, tissue loss, or major amputation if they met our EHR-based definition for PAD and possessed at least 1 diagnosis code depicted in Supplementary Table 25. If an individual possessed diagnosis codes for more than 1 severe PAD presentation (e.g. claudication and rest pain), the most severe PAD classification was selected. We then evaluated for evidence of an interaction between smoking and F5 p.R506Q carrier status.
Statistical Analysis
In our primary analysis, genotyped and imputed DNA sequence variants were tested for association with PAD using logistic regression adjusting for age, sex, and five principal components of ancestry assuming an additive model using the SNPTESTv2.5.4 (mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html) statistical software program. In our MVP discovery analysis, we performed association analyses separately for each ethnic group (whites, blacks, and Hispanics) and then meta-analyzed using an inverse variance-weighted fixed effects method implemented in the METAL software program54. We excluded variants with a high amount of heterogeneity (I2 statistic > 75%) across the three ancestries. In addition, we required that variants be observed in at least two ethnic groups.
For variants with suggestive PAD associations (P<10−6) we sought replication of our findings in UK Biobank. Association testing was performed in 5,117 PAD cases and 389,291 controls using a logistic regression model adjusted for age at baseline, sex, genotyping array, and the first 10 principal components of ancestry. All testing was performed in PLINK2 (https://www.cog-genomics.org/plink/2.0/). We defined significant novel PAD associations as those that were at least nominally significant in replication (P<0.05), were directionally consistent in both cohorts, and had an overall P < 5 ×10−8 (genome-wide significance) in the discovery and replication cohorts combined. Novel loci were defined as being greater than 500,000 base-pairs away from a known PAD genome-wide associated lead variant. Additionally, linkage disequilibrium information from the 1000 Genomes Project39 was used to determine independent variants where a locus extended beyond 500,000 base-pairs. All logistic regression values of P were two-sided.
In our ABI GWAS, DNA sequence variants were tested with linear regression using an untransformed mABI value adjusting for age, sex, and five principal components of ancestry. We performed association analyses separately for each ethnic group and then meta-analyzed the results using an inverse-variance weighted fixed effects method. A P < 5.0 × 10−8 was used to declare genome-wide significance for the continuous mABI trait.
In our PheWAS analysis, DNA sequence variants were tested using logistic regression adjusting for sex, and five principal components of ancestry against disease-free controls and declared to be significantly associated with the disease if they met a P < 5.0 × 10−8. In our CAD/LAS sensitivity analysis, risk variants identified in the primary analysis were tested for association with CAD/LAS in white MVP participants and combined with either 1) publicly available summary data of 60,801 CAD case patients and 123,504 disease-free controls in the CARDIoGRAMplusC4D consortium study24 or 2) with the transancestral LAS MEGASTROKE meta-analysis25 of 6,688 LAS cases and 454,450 controls using an inverse-variance weighted fixed effects method. Variants demonstrating at least nominal (P < 0.05) significance with CAD/LAS were then re-tested with PAD after adjusting for CAD or LAS status in MVP. Variants were declared to still be associated with PAD if they demonstrated a reduction in association signal when adjusting for CAD/LAS status but PPAD remained < 0.05. All logistic regression values of P were two-sided.
In our Factor V Leiden analysis, the Leiden variant was tested for association with each subtype of PAD (intermittent claudication, rest pain, tissue loss, major amputation), as compared to PAD free controls, through logistic regression adjusting for sex, and five principal components of ancestry. Lastly, we evaluated for evidence of a Factor V Leiden-smoking interaction by stratifying MVP participants into current smokers and former/never smokers and performed a Cochran’s Q test for interaction. In our Factor V Leiden analysis, we set a Bonferroni adjusted level of significance of P=0.05/7 tests=0.007. All values of P in the Factor V Leiden analysis were two-sided.
To determine the specificity and sensitivity values of our PAD phenotype, we performed a manual chart review and calculated the resultant values using the R-3.2.0 software (Supplementary Table 26). Sensitivity refers to the ratio of (true positives)/(true positives + false negatives) and specificity the ratio of (true negatives)/(true negatives + false positives).
Natural Language Processing Algorithm for ABI
ABI values measured for patients in the VA Healthcare System are not recorded in a structured format in the EHR. Instead, the values can be found in clinical reports in narrative or semi-structured format (Extended Data Fig. 10). In order to make these ABI values available for the PAD phenotype definition, we developed a natural language processing system to identify instances of ABI values recorded within clinical notes. The system was developed in several stages and the results of an initial iteration of the system development were reported previously (abstract by Alba PR, et al, Ankle Brachial Index Extraction System. In: AMIA Annu Symp Proc; 2018). To develop a rule-based natural language processing system that could scale to process the 6.5 million documents associated with the 31,307 patients in the discovery analysis cohort, we utilized the Leo framework55, which builds on the Unstructured Information Management Architecture - Asynchronous Scaleout56. The system achieved 96.4% precision as validated on 1000 manually labeled clinical notes. A sensitivity analysis showed 89.8% recall on an instance level across 200 documents selected from the same day as a PAD diagnosis code.
Extended Data
Extended Data Figure 1 -.
Distribution of minimum ankle-brachial index values in the Million Veteran Program
Histogram of minimum ankle-brachial index (ABI) values extracted from the electronic health record for 17,861 participants of the Million Veteran Program. These values, restricted to those with an minimum ABI of < 1.4, were used for the subsequent ABI genome-wide association study.
Extended Data Figure 2 -.
Quantile-quantile plot for the discovery trans-ethnic PAD GWAS in MVP
The expected logistic regression association P values versus the observed distribution of P values for PAD association are displayed. Quantile-quantile plots were inspected for ancestry-specific analyses, and genomic control values were < 1.20 for each racial group (data not shown). No systemic inflation was observed (λgc = 1.05). All P values were two-sided. Abbreviations: PAD, Peripheral Artery Disease; GWAS, Genome-wide Association Study; MVP, Million Veteran Program
Extended Data Figure 3 -.
Manhattan plot for the PAD GWAS
Plot of -log10(P) for association of imputed variants by chromosomal position for all autosomal polymorphisms analyzed in the PAD GWAS. The genes nearest to the top associated variants are displayed. Genes highlighted in red represent novel PAD loci (18). Genes for variants that are outside the transcript boundary of a protein-coding gene are shown with nearest candidate gene in parentheses [eg, (LDLR)]. Logistic regression two-sided P values are displayed.
Abbreviations: PAD, Peripheral artery disease; GWAS, genome-wide association study
Extended Data Figure 4 -.
TCF7L2 mediates its effect on PAD via type 2 diabetes
a) Forest plot depicting the replication of the known TCF7L2/rs7903146-T2D association signal in MVP for both white and black participants. b) The same variant is also associated with PAD risk in whites and blacks in MVP. However, when controlling for T2D status in the regression model, c) the association signal is dramatically reduced suggesting that TCF7L2 PAD risk is mediated through its effect on T2D. Logistic regression two-sided values of P are displayed.
Abbreviations: MVP, Million Veteran Program; PAD, Peripheral Artery Disease; T2D, Type 2 Diabetes
Extended Data Figure 5 -.
Forest plot for association of the CHRNA3 locus and peripheral artery disease risk stratified by smoking status
When stratifying European MVP participants by smoking status (ever smokers vs. never smokers), nearly all the association signal resides within the ever smoker group. Previous reports of variation at the CHRNA3 locus demonstrate that carriers of the PAD risk allele have a reduced likelihood of cigarette smoking cessation1. This suggests that the PAD-CHRNA3 association is driven by a greater burden of tobacco exposure in those who carry the nicotine dependence/PAD risk allele. Logistic regression two-sided values of P are displayed.
Abbreviations: MVP, Million Veteran Program; PAD, Peripheral Artery Disease
Extended Data Figure 6 -.
Peripheral artery disease risk loci and known causal risk factors
Peripheral artery disease risk loci identified in this GWAS analysis are depicted along with the plausible relationship to the underling causal risk factor. Loci names are based on the nearest genes; however, the causal gene(s) remains unclear for some associated loci and as such, the resultant annotation may prove incorrect in some cases.
Abbreviations: GWAS, Genome-wide Association Study
Extended Data Figure 7 -.
Peripheral artery disease risk variants and association with LAS and CAD
For the 19 PAD risk variants identified in our study, logistic regression Z-scores of association (aligned to the PAD risk allele) were obtained from MVP and publicly available summary statistics for large artery stroke (MVP + MEGASTROKE consortium2) and coronary artery disease (MVP + CARDIoGRAMplusC4D consortium3). A positive Z-score (red) indicates a positive association between the PAD risk allele and the disease, while a negative Z-score (blue) indicates an inverse association. Boxes are outlined in cyan if the variant is uniquely associated with PAD (two-sided PPAD < 5 ×10−8, PCAD & PLAS > 0.05).
Abbreviations: PAD, Peripheral Artery Disease; LAS, Large Artery Stroke; CAD, Coronary Artery Disease
Extended Data Figure 8 -.
Peripheral artery disease risk variants and mechanistic overlap with LAS and CAD
Venn diagram of each of the 19 PAD risk loci in a based on their association with PAD (two-sided PPAD < 5 ×10−8), CAD (P < 0.05), and LAS (P < 0.05). Each locus is depicted along with the plausible relationship to the underling causal risk factor separately by color. Loci names are based on the nearest genes; however, the causal gene(s) remains unclear for some associated loci and as such, the resultant annotation may prove incorrect in some cases.
Abbreviations: PAD, Peripheral Artery Disease; LAS, Large Artery Stroke; CAD, Coronary Artery Disease
Extended Data Figure 9 -.
Overall study design
The primary analysis consisted of a genome-wide association study to identify novel PAD risk variants. Secondary analyses involved a genome-wide association study of minimum ABI, a closer examination the 19 PAD risk variants through PheWAS, a candidate causal gene analysis using eQTL/pQTL/TWAS data, a PAD analysis accounting for CAD/LAS status, and a focused Factor V Leiden analysis.
Abbreviations: MVP, Million Veteran Program; PAD, Peripheral artery disease; ABI, Ankle-Brachial Index; CAD, Coronary Artery Disease; LAS, Large Artery Stroke; PheWAS, Phenome-wide Association Study
Extended Data Figure 10 -.
Natural Language Processing for index extraction
Examples of semi-structured text that contains targeted indices for extraction using Natural Language Processing (NLP)
Abbreviations: ABI, Ankle-Brachial Index; TBI, Toe-Brachial Index; PT, Posterior Tibial Artery; AT, Anterior Tibial Artery
Supplementary Material
Acknowledgments:
This research is based on data from the Million Veteran Program, Office of Research and Development, Veterans Health Administration, and was supported by award #MVP000. This publication does not represent the views of the Department of Veterans Affairs or the United States Government. This research was also supported by funding from: the Department of Veterans Affairs awards I01–01BX03340 (K.C., P.W.F.W.), I01-BX003362 (P.S.T., K.M.C), and IK2-CX001780 (S.M.D.) and the VA Informatics and Computing Infrastructure (VINCI) VA HSR RES 130457 (P.A., O.V.P., S.L.D.); the National Institutes of Health grants R01-HL131977 (J.A.B.), 1R03AG050930 (S.A.), R01-HL138306 (J.C.), R01HL127564 (S.K.) and K08HL140203 (P.N.); and the American Heart Association grant 18SFRN33960373 (J.A.B, M.S.F), 17IFUNP33840012 (K.G.A), and 15MCPRP25580005 (S.A.). Data on coronary artery disease have been contributed by the CARDIoGRAMplusC4D investigators. Data on large artery stroke have been contributed by the MEGASTROKE investigators. The MEGASTROKE project received funding from sources specified at http://www.megastroke.org/acknowledgements.html.
Competing Interests
J.A.B. reports consulting with Astra Zeneca, Bristol Myers Squibb, Amgen, Merck, Sanofi, Antidote Pharmaceutical, and Boehringer Ingelheim. He serves on the DSMC for Bayer and Novartis. S.L.D. has received research grant support from the following for-profit companies through the University of Utah or the Western Institute for Biomedical Research (VA Salt Lake City’s affiliated non-profit): AbbVie Inc., Anolinx LLC, Astellas Pharma Inc., AstraZeneca Pharmaceuticals LP, Boehringer Ingelheim International GmbH, Celgene Corporation, Eli Lilly and Company, Genentech Inc., Genomic Health, Inc., Gilead Sciences Inc., GlaxoSmithKline PLC, Innocrin Pharmaceuticals Inc., Janssen Pharmaceuticals, Inc., Kantar Health, Myriad Genetic Laboratories, Inc., Novartis International AG, and PAREXEL International Corporation. S.M.D. receives research support to his institution from CytoVAS and RenalytixAI. S.K. is a founder of Maze Therapeutics, Verve Therapeutics, and San Therapeutics. He holds equity in Catabasis and San Therapeutics. He is a member of the scientific advisory boards for Regeneron Genetics Center and Corvidia Therapeutics; served as a consultant for Acceleron, Eli Lilly,Novartis,Merck,NovoNordisk,Novo Ventures, Ionis, Alnylam, Aegerion, Huag Partners, Noble Insights, Leerink Partners, Bayer Healthcare, Illumina, Color Genomics, MedGenome, Quest, and Medscape; and reports patents related to a method of identifying and treating a person having a predisposition to or afflicted with cardiometabolic disease (20180010185) and a genetics risk predictor (20190017119). O.V.P. has received research grants from the following for-profit organizations through the University of Utah or Western Institute for Biomedical Research: Anolinx LLC, AstraZeneca Pharmaceuticals LP, Genentech Inc., Genomic Health, Inc., Gilead Sciences Inc., Janssen Pharmaceuticals, Inc., Novartis International AG, and PAREXEL International Corporation.
Footnotes
Data availability
The full summary level association data from the MVP trans-ancestry PAD meta-analysis from this report are available through dbGAP, accession code phs001672.v2.p1. Additional data that support the findings of this study are available on request from the corresponding author (S.M.D.); these data are not publicly available due to U.S. Government and Department of Veteran’s Affairs restrictions relating to participant privacy and consent. Data contributed by CARDIoGRAMplusC4D investigators are available online (http://www.CARDIOGRAMPLUSC4D.org/). Data on large artery stroke have been contributed by the MEGASTROKE investigators and are available online (http://www.megastroke.org/). The genetic and phenotypic UK Biobank data are available upon application to the UK Biobank. Source data has been provided for Figures 2, 3, and Extended Data Figures 4, 5, and 7. Additional data that were used to generate the Figures in this study are available on request from the corresponding author (S.M.D.) or through dbGAP as listed above.
Code availability
Code to perform analyses in this manuscript are available from the authors upon request (D.K., S.M.D.), or from the URLs associated with each software in the online methods section.
References
- 1.GBD 2016 Causes of Death Collaborators. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 390, 1151–1210 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wahlgren CM & Magnusson PK Genetic influences on peripheral arterial disease in a twin population. Arterioscler Thromb Vasc Biol 31, 678–82 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Murabito JM et al. Association between chromosome 9p21 variants and the ankle-brachial index identified by a meta-analysis of 21 genome-wide association studies. Circ Cardiovasc Genet 5, 100–12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsukura M et al. Genome-Wide Association Study of Peripheral Arterial Disease in a Japanese Population. PLoS One 10, e0139262 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins R What makes UK Biobank special? The Lancet 379, 1173–1174 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Gaziano JM et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol 70, 214–23 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Fan J et al. Billing code algorithms to identify cases of peripheral artery disease from administrative data. J Am Med Inform Assoc 20, e349–54 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kullo IJ et al. The ATXN2-SH2B3 locus is associated with peripheral arterial disease: an electronic medical record-based genome-wide association study. Front Genet 5, 166 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thorgeirsson TE et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature 452, 638–642 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denny JC et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics 26, 1205–10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denny JC et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol 31, 1102–10 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voight BF et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 42, 579–89 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klarin D, Emdin CA, Natarajan P, Conrad MF & Kathiresan S Genetic Analysis of Venous Thromboembolism in UK Biobank Identifies the ZFPM2 Locus and Implicates Obesity as a Causal Risk Factor. Circ Cardiovasc Genet 10(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teslovich TM et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–13 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Bakker PI et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet 38, 1166–72 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conte MS et al. Society for Vascular Surgery practice guidelines for atherosclerotic occlusive disease of the lower extremities: management of asymptomatic disease and claudication. J Vasc Surg 61, 2s–41s (2015). [DOI] [PubMed] [Google Scholar]
- 17.Staley JR et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics 32, 3207–3209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Consortium GTEx. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Angelantonio E et al. Efficiency and safety of varying the frequency of whole blood donation (INTERVAL): a randomised trial of 45 000 donors. Lancet 390, 2360–2371 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun BB et al. Genomic atlas of the human plasma proteome. Nature 558, 73–79 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gusev A et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet 48, 245–52 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anttila V et al. Analysis of shared heritability in common disorders of the brain. Science 360(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bulik-Sullivan B et al. An atlas of genetic correlations across human diseases and traits. Nat Genet 47, 1236–41 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.CARDIoGRAMplusC4D Consortium. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet 47, 1121–30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malik R et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sibon I et al. COL4A1 mutation in Axenfeld-Rieger anomaly with leukoencephalopathy and stroke. Ann Neurol 62, 177–84 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Greengard JS, Eichinger S, Griffin JH & Bauer KA Brief report: variability of thrombosis among homozygous siblings with resistance to activated protein C due to an Arg-->Gln mutation in the gene for factor V. N Engl J Med 331, 1559–62 (1994). [DOI] [PubMed] [Google Scholar]
- 28.Bertina RM et al. Mutation in blood coagulation factor V associated with resistance to activated protein C. Nature 369, 64–7 (1994). [DOI] [PubMed] [Google Scholar]
- 29.Holst AG, Jensen G & Prescott E Risk factors for venous thromboembolism: results from the Copenhagen City Heart Study. Circulation 121, 1896–903 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Cheng YJ et al. Current and former smoking and risk for venous thromboembolism: a systematic review and meta-analysis. PLoS Med 10, e1001515 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willey J et al. Epidemiology of lower extremity peripheral artery disease in veterans. J Vasc Surg (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fowkes FG et al. Comparison of global estimates of prevalence and risk factors for peripheral artery disease in 2000 and 2010: a systematic review and analysis. Lancet 382, 1329–40 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Myocardial Infarction Genetics and CARDIoGRAM Exome Consortia Investigators. Coding Variation in ANGPTL4, LPL, and SVEP1 and the Risk of Coronary Disease. N Engl J Med 374, 1134–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Musunuru K et al. Exome Sequencing, ANGPTL3 Mutations, and Familial Combined Hypolipidemia New England Journal of Medicine (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dewey FE et al. Genetic and Pharmacologic Inactivation of ANGPTL3 and Cardiovascular Disease. N Engl J Med (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emdin CA et al. Phenotypic Characterization of Genetically Lowered Human Lipoprotein(a) Levels. J Am Coll Cardiol 68, 2761–2772 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swanberg M et al. MHC2TA is associated with differential MHC molecule expression and susceptibility to rheumatoid arthritis, multiple sclerosis and myocardial infarction. Nat Genet 37, 486–94 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Anand SS et al. Rivaroxaban with or without aspirin in patients with stable peripheral or carotid artery disease: an international, randomised, double-blind, placebo-controlled trial. Lancet (2017). [DOI] [PubMed] [Google Scholar]
- 39.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loh PR, Palamara PF & Price AL Fast and accurate long-range phasing in a UK Biobank cohort. Nat Genet 48, 811–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Howie B, Fuchsberger C, Stephens M, Marchini J & Abecasis GR Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 44, 955–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price AL et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38, 904–9 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Alexander DH, Novembre J & Lange K Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19, 1655–64 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klarin D et al. Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nature Genetics (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manichaikul A et al. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–73 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winkler TW et al. Quality control and conduct of genome-wide association meta-analyses. Nat Protoc 9, 1192–212 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hyde CL et al. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet 48, 1031–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bycroft C et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bellenguez C, Strange A, Freeman C, Donnelly P & Spencer CC A robust clustering algorithm for identifying problematic samples in genome-wide association studies. Bioinformatics 28, 134–5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arya S et al. Race and Socioeconomic Status Independently Affect Risk of Major Amputation in Peripheral Artery Disease. J Am Heart Assoc 7(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carroll RJ, Bastarache L & Denny JC R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics 30, 2375–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dewey FE et al. Genetic and Pharmacologic Inactivation of ANGPTL3 and Cardiovascular Disease. N Engl J Med (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou X, Carbonetto P & Stephens M Polygenic modeling with bayesian sparse linear mixed models. PLoS Genet 9, e1003264 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willer CJ, Li Y & Abecasis GR METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cornia R, Patterson OV, Ginter T & DuVall SL. Rapid NLP Development with Leo . In: AMIA Annu Symp Proc. 2014:1356. [Google Scholar]
- 56.Ferrucci D & Lally A. UIMA: an architectural approach to unstructured information processing in the corporate research environment. Nat Lang Eng 2004;10(3–4):327–348. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.