Abstract
Understanding adaptive immunity to SARS-CoV-2 is important for vaccine development, interpreting coronavirus disease 2019 (COVID-19) pathogenesis, and calibration of pandemic control measures. Using HLA class I and II predicted peptide “megapools,” circulating SARS-CoV-2-specific CD8+ and CD4+ T cells were identified in ∼70% and 100% of COVID-19 convalescent patients, respectively. CD4+ T cell responses to spike, the main target of most vaccine efforts, were robust and correlated with the magnitude of the anti-SARS-CoV-2 IgG and IgA titers. The M, spike, and N proteins each accounted for 11%–27% of the total CD4+ response, with additional responses commonly targeting nsp3, nsp4, ORF3a, and ORF8, among others. For CD8+ T cells, spike and M were recognized, with at least eight SARS-CoV-2 ORFs targeted. Importantly, we detected SARS-CoV-2-reactive CD4+ T cells in ∼40%–60% of unexposed individuals, suggesting cross-reactive T cell recognition between circulating “common cold” coronaviruses and SARS-CoV-2.
Keywords: COVID-19, CD4, CD8, T cells, epitopes, coronavirus, SARS-CoV-2, cross-reactivity
Graphical Abstract
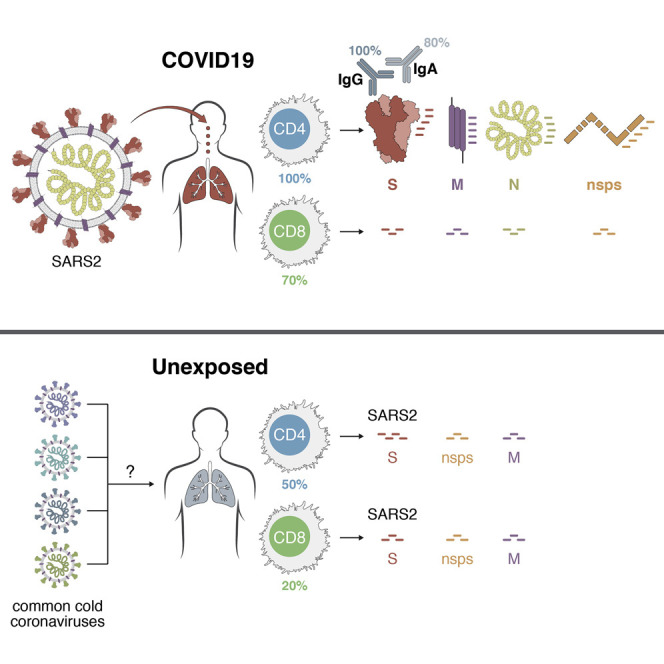
An analysis of immune cell responses to SARS-CoV-2 from recovered patients identifies the regions of the virus that is targeted and also reveals cross-reactivity with other common circulating coronaviruses
Introduction
COVID-19 is a worldwide emergency. The first cases occurred in December 2019, and now more than 240,000 deaths and 3,000,000 cases of SARS-CoV-2 infection have been reported worldwide as of May 1st (Dong et al., 2020, Wu and McGoogan, 2020). Vaccines against SARS-CoV-2 are just beginning development (Amanat and Krammer, 2020, Thanh Le et al., 2020). An understanding of human T cell responses to SARS-CoV2 is lacking, due to the rapid emergence of the pandemic. There is an urgent need for foundational information about T cell responses to this virus.
The first steps for such an understanding are the ability to quantify the virus-specific CD4+ and CD8+ T cells. Such knowledge is of immediate relevance, as it will provide insights into immunity and pathogenesis of SARS-CoV-2 infection, and the same knowledge will assist vaccine design and evaluation of candidate vaccines. Estimations of immunity are also central to epidemiological model calibration of future social distancing pandemic control measures (Kissler et al., 2020). Such projections are dramatically different depending on whether SARS-CoV-2 infection creates substantial immunity, and whether any cross-reactive immunity exists between SARS-CoV-2 and circulating seasonal “common cold” human coronaviruses. Definition and assessment of human antigen-specific SARS-CoV2 T cell responses are best made with direct ex vivo T cell assays using broad-based epitope pools and assays capable of detecting T cells of any cytokine polarization. Herein, we have completed such an assessment with blood samples from COVID-19 patients.
There is also great uncertainty about whether adaptive immune responses to SARS-CoV-2 are protective or pathogenic, or whether both scenarios can occur depending on timing, composition, or magnitude of the adaptive immune response. Hypotheses range the full gamut (Peeples, 2020), based on available clinical data from severe acute respiratory disease syndrome (SARS) or Middle East respiratory syndrome (MERS) (Alshukairi et al., 2018, Wong et al., 2004, Zhao et al., 2017) or animal model data with SARS in mice (Zhao et al., 2009, Zhao et al., 2010, Zhao et al., 2016), SARS in non-human primates (NHPs) (Liu et al., 2019, Takano et al., 2008) or feline infectious peritonitis virus (FIPV) in cats (Vennema et al., 1990). Protective immunity, immunopathogenesis, and vaccine development for COVID-19 are each briefly discussed below, related to introducing the importance of defining T cell responses to SARS-CoV-2.
Based on data from SARS patients in 2003–2004 (caused by SARS-CoV, the most closely related human betacoronavirus to SARS-CoV-2), and based on the fact that most acute viral infections result in development of protective immunity (Sallusto et al., 2010), a likely possibility has been that substantial CD4+ T cell, CD8+ T cell, and neutralizing antibody responses develop to SARS-CoV-2, and all contribute to clearance of the acute infection, and, as a corollary, some of the T and B cells are retained long term (i.e., multiple years) as immunological memory and protective immunity against SARS-CoV-2 infection (Guo et al., 2020b, Li et al., 2008). However, a contrarian viewpoint is also legitimate. While most acute infections result in the development of protective immunity, available data for human coronaviruses suggest the possibility that substantive adaptive immune responses can fail to occur (Choe et al., 2017, Okba et al., 2019, Zhao et al., 2017) and robust protective immunity can fail to develop (Callow et al., 1990). A failure to develop protective immunity could occur due to a T cell and/or antibody response of insufficient magnitude or durability, with the neutralizing antibody response being dependent on the CD4+ T cell response (Crotty, 2019, Zhao et al., 2016). Thus, there is urgent need to understand the magnitude and composition of the human CD4+ and CD8+ T cell responses to SARS-CoV-2. If natural infection with SARS-CoV-2 elicits potent CD4+ and CD8+ T cell responses commonly associated with protective antiviral immunity, COVID-19 is a strong candidate for rapid vaccine development.
Immunopathogenesis in COVID-19 is a serious concern (Cao, 2020, Peeples, 2020). It is most likely that an early CD4+ and CD8+ T cell response against SARS-CoV-2 is protective, but an early response is difficult to generate because of efficient innate immune evasion mechanisms of SARS-CoV-2 in humans (Blanco-Melo et al., 2020). Immune evasion by SARS-CoV-2 is likely exacerbated by reduced myeloid cell antigen-presenting cell (APC) function or availability in the elderly (Zhao et al., 2011). In such cases, it is conceivable that late T cell responses may instead amplify pathogenic inflammatory outcomes in the presence of sustained high viral loads in the lungs, by multiple hypothetical possible mechanisms (Guo et al., 2020a, Li et al., 2008, Liu et al., 2019). Critical (ICU) and fatal COVID-19 (and SARS) outcomes are associated with elevated levels of inflammatory cytokines and chemokines, including interleukin-6 (IL-6) (Giamarellos-Bourboulis et al., 2020, Wong et al., 2004, Zhou et al., 2020)
Vaccine development against acute viral infections classically focuses on vaccine-elicited recapitulation of the type of protective immune response elicited by natural infection. Such foundational knowledge is currently missing for COVID-19, including how the balance and the phenotypes of responding cells vary as a function of disease course and severity. Such knowledge can guide selection of vaccine strategies most likely to elicit protective immunity against SARS-CoV-2. Furthermore, knowledge of the T cell responses to COVID-19 can guide selection of appropriate immunological endpoints for COVID-19 candidate vaccine clinical trials, which are already starting.
Limited information is also available about which SARS-CoV-2 proteins are recognized by human T cell immune responses. In some infections, T cell responses are strongly biased toward certain viral proteins, and the targets can vary substantially between CD4+ and CD8+ T cells (Moutaftsi et al., 2010, Tian et al., 2019). Knowledge of SARS-CoV-2 proteins and epitopes recognized by human T cell responses is of immediate relevance, as it will allow for monitoring of COVID-19 immune responses in laboratories worldwide. Epitope knowledge will also assist candidate vaccine design and facilitate evaluation of vaccine candidate immunogenicity. Almost all of the current COVID-19 vaccine candidates are focused on the spike protein.
A final key issue to consider in the study of SARS-CoV-2 immunity is whether some degree of cross-reactive coronavirus immunity exists in a fraction of the human population, and whether this might influence susceptibility to COVID-19 disease. This issue is also relevant for vaccine development, as cross-reactive immunity could influence responsiveness to candidate vaccines (Andrews et al., 2015).
In sum, the ability to measure and understand the human CD4+ and CD8+ T cell responses to SARS-CoV-2 infection is a major knowledge gap currently impeding COVID-19 vaccine development, interpretation of COVID-19 disease pathogenesis, and calibration of future social distancing pandemic control measures.
Results
SARS-CoV-2 Peptides and Predicted Class I and Class II Epitopes
We recently predicted SARS-CoV-2 T cell epitopes utilizing the Immune Epitope Database and Analysis Resource (IEDB) (Dhanda et al., 2019, Vita et al., 2019). Utilizing bioinformatic approaches, we identified specific peptides in SARS-CoV-2 with increased probability of being T cell targets (Grifoni et al., 2020). We previously developed the megapool (MP) approach to allow simultaneous testing of large numbers of epitopes. By this technique, numerous epitopes are solubilized, pooled, and re-lyophilized to avoid cell toxicity problems (Carrasco Pro et al., 2015). These MPs have been used in human T cell studies of a number of indications, including allergies (Hinz et al., 2016), tuberculosis (Lindestam Arlehamn et al., 2016), tetanus, pertussis (Bancroft et al., 2016, da Silva Antunes et al., 2017), and dengue virus, for both CD4+ and CD8+ T cell epitopes (Grifoni et al., 2017, Weiskopf et al., 2015). Here, we generated MPs based on predicted SARS-CoV-2 epitopes. Specifically, one MP corresponds to 221 predicted HLA class II CD4+ T cell epitopes (Grifoni et al., 2020) covering all proteins in the viral genome, apart from the spike (S) antigen (CD4_R MP). The prediction strategy utilized is geared to capture ∼50% of the total response (Dhanda et al., 2018, Paul et al., 2015) and was designed and validated to predict dominant epitopes independently of ethnicity and HLA polymorphism. This approach takes advantage of the extensive cross-reactivity and repertoire overlap between different HLA class II loci and allelic variants to predict promiscuous epitopes, capable of binding many of the most common HLA class II prototypic specificities (Greenbaum et al., 2011, O’Sullivan et al., 1991, Sidney et al., 2010a, Sidney et al., 2010b, Southwood et al., 1998).
For the spike protein, to ensure that all T cell reactivity against this important antigen can be detected, we generated a separate MP covering the entire antigen with 253 15-mer peptides overlapping by 10-residues (MP_S, Table S1). As stated above, the MP used to probe the non-spike regions is expected to capture ∼50% of the total response. The use of overlapping peptides spanning entire open reading frames (ORFs) instead allows for a more complete characterization but also requires more cells. This factor should be kept in mind in terms of comparison of the magnitude of the CD4+ T cell responses to those pools.
In the case of CD8 epitopes, since the overlap between different HLA class I allelic variants and loci is more limited to specific groups of alleles, or supertypes (Sidney et al., 2008), we targeted a set of the 12 most prominent HLA class I A and B alleles, which together allow broad coverage (>85%) of the general population. Two class I MPs were synthesized based on epitope predictions for those 12 most common HLA A and B alleles (Grifoni et al., 2020), which collectively encompass 628 predicted HLA class I CD8+ T cell epitopes from the entire SARS-CoV-2 proteome (CD8 MP-A and MP-B).
Immunological Phenotypes of Recovered COVID-19 Patients
To test for the generation of SARS-CoV-2 CD4+ and CD8+ T cell responses following infection, we initially recruited 20 adult patients who had recovered from COVID-19 disease (Table 1 ). We also utilized peripheral blood mononuclear cell (PBMC) and plasma samples from local healthy control donors collected in 2015–2018 (see STAR Methods). Blood samples were collected at 20–35 days post-symptoms onset from non-hospitalized COVID-19 patients who were no longer symptomatic. SARS-CoV-2 infection was determined by swab test viral PCR during the acute phase of the infection. Verification of SARS-CoV-2 exposure was attempted both by lateral flow serology and SARS-CoV-2 spike protein receptor binding domain (RBD) ELISA (Stadlbauer et al., 2020), using plasma from the convalescence stage blood draw. Most patients were confirmed positive by lateral flow immunoglobulin (Ig) tests (Table 1). All patients were confirmed COVID-19 cases by SARS-CoV-2 RBD ELISA (Figures 1 and S1 ). All cases were IgG positive; anti-RBD IgM and IgA was also detected in the large majority of cases (Figures 1 and S1).
Table 1.
Participant Characteristics
| Age (years) | Unexposed (n = 20) |
COVID-19 (n = 20) |
|---|---|---|
| 20–66 (median = 31, IQR = 21) | 20–64 (median = 44, IQR = 9) | |
| Gender | ||
| Male (%) | 35% (7/20) | 45% (9/20) |
| Female (%) | 65% (13/20) | 55% (11/20) |
| Residency | ||
| California (%) | 95% (19/20) | 100% (20/20) |
| USA, Non-California (%) | 5% (1/20) | 0% (0/20) |
| Sample Collection Date | March 2015–March 2018 | March–April 2020 |
| SARS-CoV-2 PCR Positivity | N/A | 100% (16/16 tested) |
| Antibody Test Positivitya | N/A | 90% (18/20) |
| Disease Severityb | ||
| Mild | N/A | 70% (14/20) |
| Moderate | N/A | 20% (4/20) |
| Severe | N/A | 10% (2/20) |
| Critical | N/A | 0% (0/20) |
| Symptoms | ||
| Cough | N/A | 79% (15/19) |
| Fatigue | N/A | 42% (8/19) |
| Fever | N/A | 37% (7/19) |
| Anosmia | N/A | 21% (4/19) |
| Dyspnea | N/A | 16% (3/19) |
| Diarrhea | N/A | 5% (1/19) |
| Days Post Symptom Onset at Collection | N/A | 20–36 (18/20) (median = 26, IQR = 7) |
| Past Medical History | ||
| No known | N/A | 65% (13/20) |
| Hyperlipidemia | N/A | 15% (3/20) |
| Hypertension | N/A | 10% (2/20) |
| Asthma | N/A | 10% (2/20) |
| Known or suspected sick contact/exposure | N/A | 75% (15/20) |
Commercial skin prick lateral flow assay.
WHO criteria.
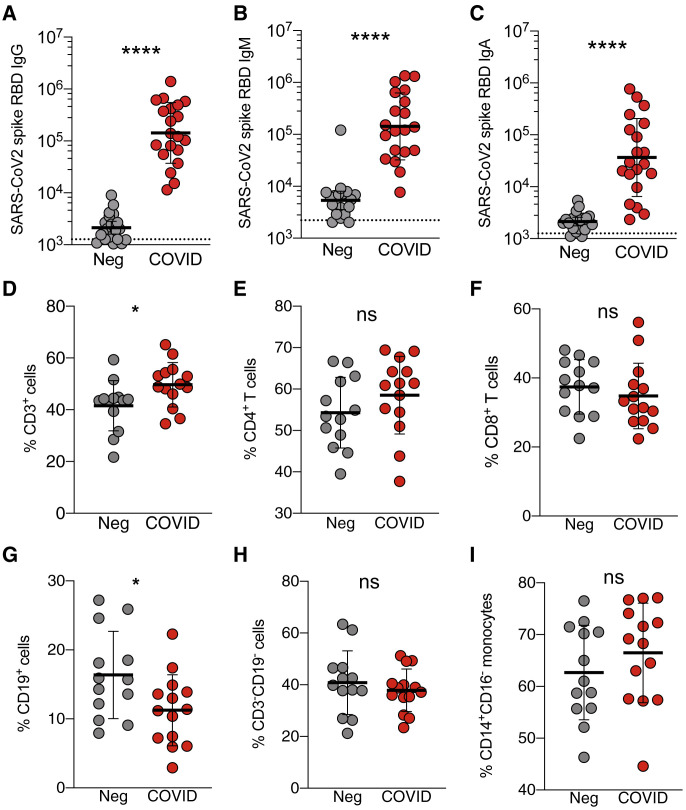
Figure 1.
SARS-CoV-2 IgM, IgA, and IgG Responses of Recovered COVID-19 Patients
(A–C) Plasma ELISA titers to SARS-CoV-2 spike RBD. (A) IgG. (B) IgM. (C) IgA. Neg, unexposed donors from 2015–2018 (n = 20); COVID, convalescing COVID-19 patients (n = 20). All data are shown as ELISA titers based on a standard. The dotted line indicates limit of detection. Geometric mean titers with geometric SDs are indicated.
(D–I) Immunophenotyping of mononuclear leukocytes. Frequency of (D) CD3+ total T cells, (E) CD4+ T cells (CD4+CD3+), (F) CD8+ T cells (CD8+CD3+), (G) CD19+ B cells (CD19+CD3–), (H) CD3–CD19– cells, and (I) CD14+CD16– monocytes (CD3–CD19–CD56–) from the PBMCs of unexposed donors (Neg, n = 13) or convalescing COVID-19 patients (COVID, n = 14). Data were analyzed using the Mann-Whitney test with mean and standard deviation shown.
∗p < 0.05, ∗∗∗∗p < 0.0001. See also Figures S1 and S2.
Figure S1.
SARS-CoV-2 Spike Protein RBD Serology, Related to Figure 1
(A-C) ELISA curves for (A) IgG, (B) IgM, and (C) IgA from 10 representative donors. Five COVID-19 cases and
(D-F) Area under the curve (AUC) SARS-CoV-2 spike protein RBD (D) IgG (E) IgM, and (F) IgA, ELISA quantitation, from the same donors and experiments shown in Figure 1. Geometric mean titers with geometric SDs are indicated. P values are two-tailed Mann-Whitney tests.
We defined a 21-color flow cytometry panel of mononuclear leukocyte lineage and phenotypic markers (Table S2) to broadly assess the immunological cellular profile of recovered COVID-19 patients (Figures 1 and S2 ). The frequency of CD3+ cells was slightly increased in recovered COVID-19 patients relative to non-exposed controls, while no significant differences overall were observed in the frequencies of CD4+ or CD8+ T cells between the two groups. Frequencies of CD19+ cells were somewhat decreased, while no differences were observed in the frequencies of CD3–CD19– cells or CD14+CD16– monocytes (Figures 1 and S2). No evidence of general lymphopenia was observed in the convalescing patients, consistent with the literature. Next, we utilized the SARS-CoV-2 MPs to probe CD4+ and CD8+ T cell responses.
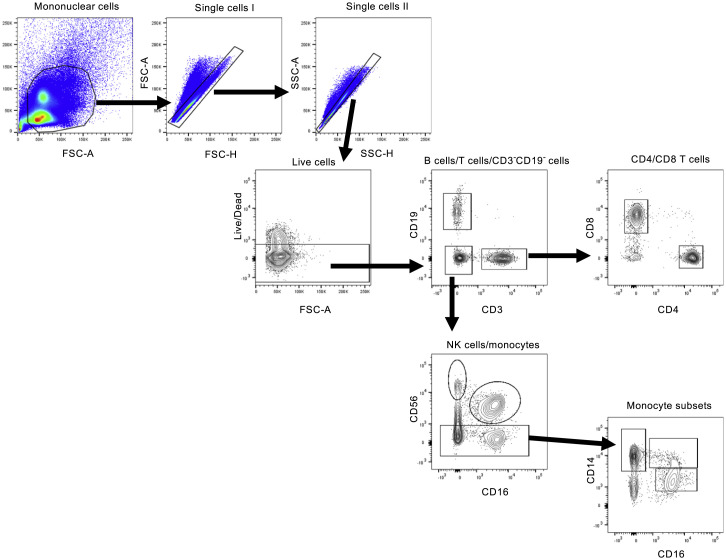
Figure S2.
Phenotyping Flow Cytometry, Related to Figure 1
Representative gating of CD3+ T cells, CD19+ B cells, CD3-CD19- cells, CD4+ T cells, CD8+ T cells and CD14+ monocytes from donor PBMCs is shown. Briefly, mononuclear cells were gated out of all events followed by subsequent singlet gating. Live cells are gated as Zombie UV-. Cells were then gated as CD19-PE-Cy5+, CD3-buv395+ or CD19-CD3- cells. T cells were further subdivided into either CD8-buv805+ or CD4-PerCPefluor710+ populations. CD3-CD19- cells were defined as CD56-PE-Dazzlebright NK cells, CD56dimCD-16buv737+ NK cells or CD56- monocytes. Monocytes were further classified on differential expression of CD14-bv510 and CD16.
Identification and Quantitation of SARS-CoV-2-Specific CD4+ T Cell Responses
We utilized T cell receptor (TCR) dependent activation induced marker (AIM) assays to identify and quantify SARS-CoV-2-specific CD4+ T cells in recovered COVID-19 patients. Initial definition and assessment of human antigen-specific SARS-CoV-2 T cell responses are best made with direct ex vivo T cell assays using broad-based epitope pools, such as MPs, and assays capable of detecting T cells of unknown cytokine polarization and functional attributes. AIM assays are cytokine-independent assays to identify antigen-specific CD4+ T cells (Havenar-Daughton et al., 2016, Reiss et al., 2017). AIM assays have been successfully used to identify virus-specific, vaccine-specific, or tuberculosis-specific CD4+ T cells in a range of studies (Dan et al., 2016, Dan et al., 2019, Herati et al., 2017, Morou et al., 2019).
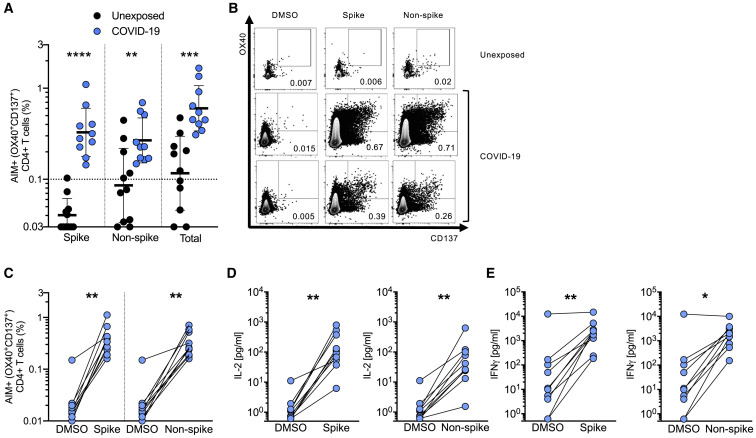
We stimulated PBMCs from 10 COVID-19 cases and 11 healthy controls (SARS-CoV-2 unexposed, collected in 2015–2018) with a spike MP (MP_S) and the class II MP covering the remainder of the SARS-CoV-2 orfeome (“non-spike,” MP CD4_R). A CMV MP was used as a positive control, while DMSO was used as the negative control (Figures 2 and S3 ). SARS-CoV-2 spike-specific CD4+ T cell responses (OX40+CD137+) were detected in 100% of COVID-19 cases (p < 0.0001 versus unexposed donors spike MP, Figures 2A and 2B. p = 0.002 versus DMSO control, Figure 2C). CD4+ T cell responses to the remainder of the SARS-CoV-2 orfeome were also detected in 100% of COVID-19 cases (p < 0.0079 versus unexposed donors non-spike MP, Figures 2A and 2B. p = 0.002, non-spike versus DMSO control, Figure 2C). The magnitude of the SARS-CoV-2-specific CD4+ T cell responses measured was similar to that of the CMV MP (Figure S3C). The concordance between SARS-CoV-2-specific CD4+ T cell measurements in independent experiments was high (p < 0.0002, Figure S3D). To assess functionality and polarization of the SARS-CoV-2-specific CD4+ T cell response, we measured cytokines secreted in response to MP stimulation. The SARS-CoV-2-specific CD4+ T cells were functional, as the cells produced IL-2 in response to non-spike and spike MPs (Figure 2D). Polarization of the cells appeared to be a classic TH1 type, as substantial interferon (IFN)-γ was produced (Figure 2E), while little to no IL-4, IL-5, IL-13, or IL-17α was expressed (Figures S3G–S3J).
Figure 2.
SARS-CoV-2-Specific CD4+ T Cell Responses of Recovered COVID-19 Patients
(A) SARS-CoV-2-specific CD4+ T cells measured as percentage of AIM+ (OX40+CD137+) CD4+ T cells after stimulation of PBMCs with peptide pools encompassing spike only (Spike) MP or the CD4_R MP representing all the proteome without spike (Non-spike). Data were background subtracted against DMSO negative control and are shown with geometric mean and geometric standard deviation. Samples were from unexposed donors (Unexposed, n = 11) and recovered COVID-19 patients (COVID-19, n = 10).
(B) Fluorescence-activated cell sorting (FACS) plot examples, gated on total CD4+ T cells.
(C) AIM+ CD4+ T cell reactivity in COVID-19 cases between the negative control (DMSO) and antigen-specific stimulations.
(D and E) Cytokine levels in the supernatant of PBMCs from COVID-19 donors after stimulation with peptide pools (Spike and Non-spike) or the negative control (DMSO). (D) IL-2. (E) IFN-γ.
Statistical comparisons across cohorts were performed with the Mann-Whitney test. Pairwise comparisons (C–E) were performed with the Wilcoxon test. ∗∗p < 0.01; ∗∗∗p < 0.001. See also Figure S3 and Table S6.
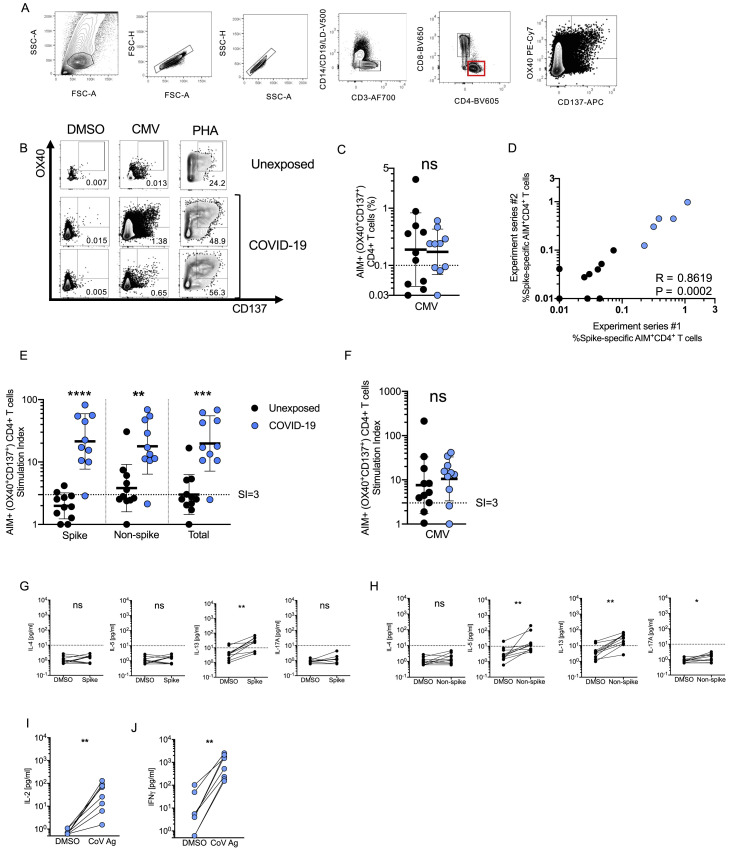
Figure S3.
SARS-CoV-2-Specific CD4+ T Cell Responses of Recovered COVID-19 Patients, Related to Figure 2
(A) Example flow cytometry gating strategy.
(B) FACS plot examples for controls. DMSO negative control, CMV positive control, PHA positive control.
(C) CMV-specific CD4+ T cells as percentage of AIM+ (OX40+CD137+) CD4+ T cells after stimulation of PBMCs with CMV peptide pool. Data were background subtracted against DMSO negative control and are shown with geometric mean and geometric standard deviation. Samples were from unexposed donors (“Unexposed,” n = 11) and recovered COVID-19 patients (“COVID-19,” n = 10).
(D) Spearman correlation of SARS-CoV-2 spike−specific CD4+ T cells (AIM+ (OX40+CD137+) CD4+ T cells, background subtracted) after stimulation with spike pool run on the same donors in two independent experiment series run on different dates. COVID-19 patient samples shown in blue. Unexposed donor samples shown in black.
(E-F) Stimulation index quantitation of AIM+ (OX40+CD137+) CD4+ T cells; the same samples as in Figure 2 and Figure S3C were analyzed.
(G-H) Cytokine levels in the supernatants of AIM assays after stimulation with (G) Spike MP (MP_S), or (H) CD4-R (“Non-spike”). Data are shown in comparison to the negative control (DMSO), per donor.
(I-J) Cytokine production by CD4+ T cells in response to Non-spike (CD4-R MP) or Spike (MP_S) peptide pools (“CoV antigen (Ag)”) was confirmed by analyzing cytokine secretion from the subset of COVID-19 donors determined to have low or negative CD8+ T cell responses (< 0.1% by AIM) to the same peptide pool determined positive for SARS-CoV-2−specific CD4+ T cells by AIM. (I) IL-2. (J) IFNγ.
Statistical comparisons across cohorts were performed with the Mann-Whitney test, while paired sample comparisons were performed with the Wilcoxon test. ∗∗p < 0.01; ∗∗∗p < 0.001. ns not significant.
Thus, recovered COVID-19 patients consistently generated a substantial CD4+ T cell response against SARS-CoV-2. Similar conclusions were reached using stimulation index as the metric (Figures S3E and S3F). In terms of total CD4+ T cell response per donor (Figure 2A), on average ∼50% of the detected response was directed against the spike protein, and ∼50% was directed against the MP representing the remainder of the SARS-CoV-2 orfeome (Figure 2A). This is of significance, since the SARS-CoV-2 spike protein is a key component of the vast majority of candidate COVID-19 vaccines under development. Of note, given the nature of the MP_R peptide predictions, the actual CD4+ T cell response to be ascribed to non-spike ORFs was likely to be higher, addressed in further experiments below.
Identification and Quantitation of SARS-CoV-2-Specific CD8+ T Cell Responses
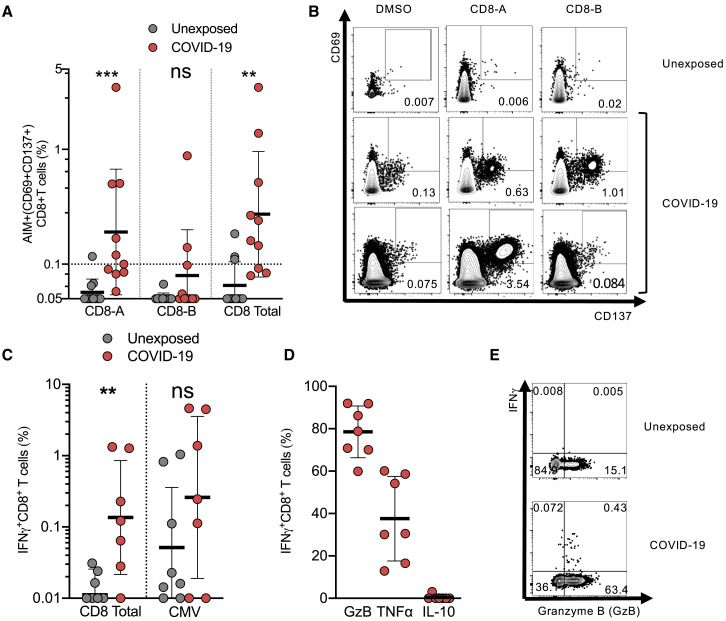
To measure SARS-CoV-2-specific CD8+ T cells in the recovered COVID-19 patients, we utilized two complementary methodologies, AIM assays and intracellular cytokine staining (ICS). The two SARS-CoV-2 class I MPs were used, CD8-A and CD8-B, with CMV MP and DMSO serving as positive and negative controls, respectively (Figures 3 and S4 ). CD8+ T cell responses were detected by AIM (CD69+CD137+) in 70% of COVID-19 cases (p < 0.0011 versus unexposed donors “CD8 total,” Figures 3A and 3B; p = 0.002, CD8-A or CD8-B versus DMSO control, Figure S4B). MP CD8-A contains spike epitopes, among epitopes to other proteins. The magnitude of the SARS-CoV-2 reactive CD8+ T cell responses measured by AIM was somewhat lower than the CMV MP (Figure S4C). Similar conclusions were reached using stimulation index (Figures S3D and S3E).
Figure 3.
SARS-CoV-2-Specific CD8+ T Cell Responses by Recovered COVID-19 Patients
(A) SARS-CoV-2-specific CD8+ T cells measured as percentage of AIM+ (CD69+CD137+) CD8+ T cells after stimulation of PBMCs with class I MPs (CD8-A, CD8-B, and the combined data [Total]). Data were background subtracted against DMSO negative control and are shown with geometric mean and geometric standard deviation. Samples were from unexposed donors (Unexposed, n = 11) and recovered COVID-19 patients (COVID-19, n = 10).
(B) FACS plot examples.
(C) Percentage of CD8+ T cells producing IFN-γ in response to SARS-CoV-2 MPs, or CMV MP, in PBMCs from COVID-19 and unexposed donors after background subtraction. Data are shown with geometric mean and geometric standard deviation.
(D) Functional profile of IFN-γ+CD8+ T cells producing granzyme B (GzB), TNF-α (TNF), or IL-10 in response to SARS-CoV-2 MPs. Mean and SD are shown.
(E) FACS plot examples of IFN-γ and granzyme B co-expression.
Statistical comparisons across cohorts were performed with the Mann-Whitney test. ∗p < 0.05; ∗∗p < 0.01.; ns not significant. See also Figure S4 and Table S6.
Figure S4.
SARS-CoV-2-Specific CD8+ T Cell Responses of Recovered COVID-19 Patients, Related to Figure 3
(A) Flow cytometry gating strategy.
(B) SARS-CoV-2−specific CD8+ T cells as determined by AIM+ (CD69+CD137+) CD8+ T cells. Response of PBMCs from COVID-19 cases between the negative control (DMSO) and antigen specific stimulation.
(C) CMV-specific CD8+ T cells as percentage of AIM+ (CD69+CD137+) CD8+ T cells after stimulation of PBMCs with CMV peptide pool. Data were background subtracted against DMSO negative control and are shown with geometric mean and geometric standard deviation. Samples were from unexposed donors (“Unexposed,” n = 11) and recovered COVID-19 patients (“COVID-19,” n = 10).
(D-E) Stimulation index quantitation of AIM+ (CD69+CD137+) CD8+ T cells; the same samples as in Figure 2 and Figure S4C were analyzed.
Statistical comparisons across cohorts were performed with the Mann-Whitney test, while paired sample comparisons were performed with the Wilcoxon test. ∗∗p < 0.01; ∗∗∗p < 0.001. ns not significant.
Independently, ICS assays detected IFN-γ+ SARS-CoV-2-specific CD8+ T cells in the majority of COVID-19 cases (Figures 3C and 3D). The majority of IFN-γ+ cells co-expressed granzyme B (Figures 3D and 3E). A substantial fraction of the IFN-γ+ cells expressed tumor necrosis factor (TNF) but not IL-10 (Figure 3D). Thus, the majority of recovered COVID-19 patients generated a CD8+ T cell response against SARS-CoV-2.
Relationship between SARS-CoV-2-Specific CD4+ T Cell Responses and IgG and IgA Titers
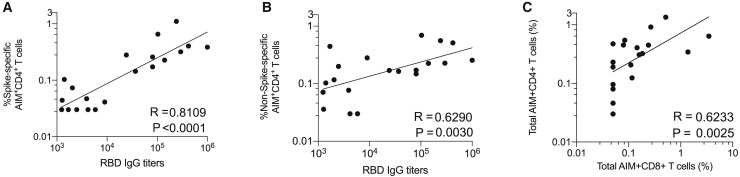
Most protective antibody responses are dependent on CD4+ T cell help. Therefore, we assessed whether stronger SARS-CoV-2-specific CD4+ T cell responses were associated with higher antibody titers in COVID-19 cases. Given that spike is the primary target of SARS neutralizing antibodies, we examined spike-specific CD4+ T cells. Spike-specific CD4+ T cell responses correlated well with the magnitude of the anti-spike RBD IgG titers (R = 0.81; p < 0.0001; Figure 4 A). Similar results were obtained using stimulation index (Figure S5 A). The non-spike SARS-CoV-2-specific CD4+ T cell response did not correlate as well with anti-spike RBD IgG titers (Figures 4B and S5B), consistent with a common requirement for intramolecular CD4+ T cell help (Sette et al., 2008). Anti-spike IgA titers also correlated with spike-specific CD4+ T cells (p < 0.0002, Figure S5). Thus, COVID-19 patients make anti-spike RBD antibody responses commensurate with the magnitude of their spike-specific CD4+ T cell response. We then assessed the relationship between the CD4+ and CD8+ T cell responses to SARS-CoV-2. SARS-CoV-2-specific CD4+ and CD8+ T cell responses were well correlated (R = 0.62. p = 0.0025, Figures 4C and S5). Thus, antibody, CD4+, and CD8+ T cell responses to SARS-CoV-2 were generally well correlated.
Figure 4.
Correlations between SARS-CoV-2-Specific CD4+ T Cells, Antibodies, and CD8+ T Cells
(A) Correlation between SARS-CoV-2 spike-specific CD4+ T cells (%) and anti-spike RBD IgG.
(B) Correlation between SARS-CoV-2 non-spike-specific CD4+ T cells (%) and anti-spike RBD IgG.
(C) Correlation between SARS-CoV-2-specific CD4+ T cells and SARS-CoV-2-specific CD8+ T cells. Total MP responses per donor were used in each case (“Non-spike” + “spike” (CD4_R + MP_S) for CD4+ T cells, CD8_A + CD8_B for CD8+ T cells).
Statistical comparisons were performed using Spearman correlation. See also Figure S5.
Figure S5.
Correlations between SARS-CoV-2-Specific CD4+ T Cells, Antibodies, and CD8+ T Cells, Related to Figure 4
(A) Correlation between SARS-CoV-2 spike−specific CD4+ T cells and anti-spike RBD IgG, using CD4+ T cell stimulation index.
(B) Correlation between SARS-CoV-2 non-spike−specific CD4+ T cells and anti-spike RBD IgG, using CD4+ T cell stimulation index.
(C) Correlation between SARS-CoV-2 spike−specific CD4+ T cells (%) and anti-spike RBD IgA.
(D) Correlation between SARS-CoV-2 spike−specific CD4+ T cells (%) and anti-spike RBD IgA.
(E) Correlation between SARS-CoV-2−specific CD4+ T cells and SARS-CoV-2−specific CD8+ T cells, using stimulation index. Total MP responses per donor were used in each case (“Non-spike” + “spike” (CD4_R + MP_S) for CD4+ T cells, CD8-A + CD8-B for CD8+ T cells).
Statistical comparisons were performed using Spearman correlation.
Pre-existing Cross-Reactive Coronavirus-Specific T Cells
While spike- and non-spike-specific CD4+ T cell responses were detectable in all COVID-19 cases, cells were also detected in unexposed individuals (Figures 3A and 3B). These responses were statistically significant for non-spike-specific CD4+ T cell reactivity (non-spike, p = 0.039; spike, p = 0.067; Figures 5A and 5B). Non-spike-specific CD4+ T cell responses were above the limit of detection in 50% of donors based on stimulation index (SI) (Figure S3E). All of the donors were recruited between 2015 and 2018, excluding any possibility of exposure to SARS-CoV-2. Four human coronaviruses are known causes of seasonal “common cold” upper-respiratory tract infections: HCoV-OC43, HCoV-HKU1, HCoV-NL63, and HCoV-229E. We tested the SARS-CoV-2 unexposed donors for seroreactivity to HCoV-OC43 and HCoV-NL63 as a representative betacoronavirus and alphacoronavirus, respectively. All donors were IgG seropositive to HCoV-OC43 and HCoV-NL63 RBD, to varying degrees (Figure 5C), consistent with the endemic nature of these viruses (Gorse et al., 2010, Huang et al., 2020, Severance et al., 2008). We therefore examined whether these represented true pan-coronavirus T cells capable of recognizing SARS-CoV-2 epitopes.
Figure 5.
SARS-CoV-2 Epitope Reactivity in Unexposed Individuals
(A) SARS-CoV-2-reactive CD4+ T cells measured as percentage of AIM+ (OX40+CD137+) CD4+ T cells in unexposed (n = 11) donors.
(B) FACS plot examples, gated on total CD4+ T cells.
(C) Plasma IgG ELISAs for seroreactivity to RBD of HCoV-OC43 or HCoV-NL63. Data are expressed as geometric mean and geometric SD.
Pairwise statistical comparisons (A) were performed with the Wilcoxon test. ∗p < 0.05; ns, not significant.
SARS-CoV-2 ORF Targets of CD4+ and CD8+ T Cells
A most pressing, yet unresolved, set of issues in understanding SARS-CoV-2 immune responses is what antigens are targeted by CD4+ and CD8+ T cells, whether the corresponding antigens are the same or different, and how do they reflect the antigens currently considered for COVID-19 vaccine development. We synthesized sets of overlapping peptides spanning the entire sequence of SARS-CoV-2 and pooled them separately so that each pool would represent one antigen (with the exception of nsp3, for which two pools were made; Table S1).
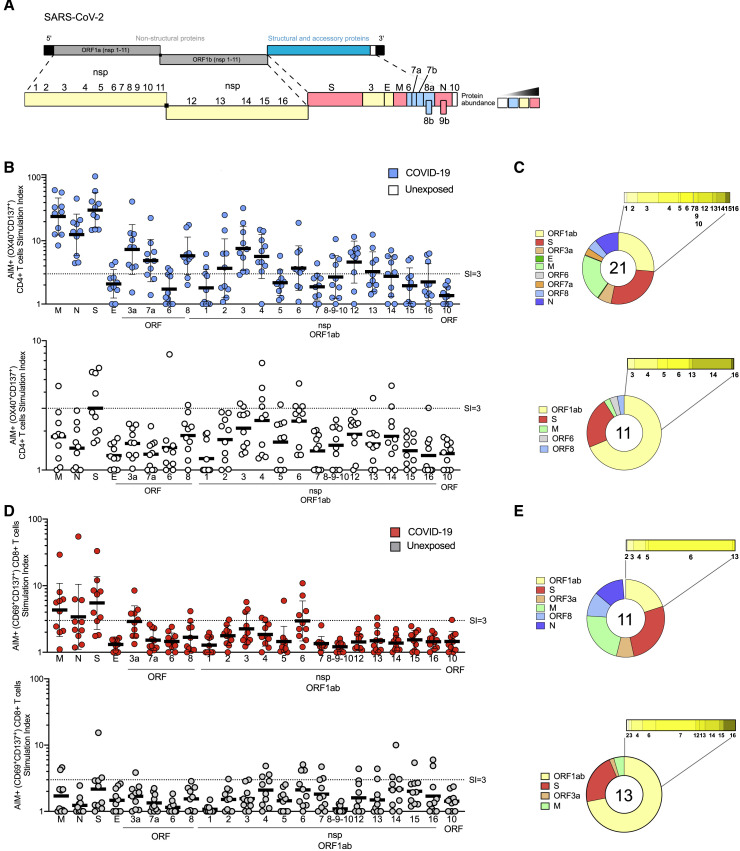
In the case of CD4+ T cell responses, no obvious pattern of antigen specificity was observed based on SARS-CoV-2 genome organization; however, coronaviruses increase protein synthesis of certain ORFs in infected cells via subgenomic RNAs. Accounting for the relative abundance of subgenomic RNAs (Figure 6 A) (Irigoyen et al., 2016, Snijder et al., 2003, Xie et al., 2020), the ORFs were re-ordered based on predicted protein abundance (Figure 6B). A clear hierarchy of SARS-CoV-2-specific CD4+ T cell targets was then apparent, with the majority of the CD4+ T cell response in COVID-19 cases directed against highly expressed SARS-CoV-2 ORFs spike, M, and N. On average, these antigens accounted for 27%, 21%, and 11% of the total CD4+ T cell response, respectively. Most COVID-19 cases also had CD4+ T cells specific for SARS-CoV-2 nsp3, nsp4, and ORF8 (Figure 6B), on average each accounting for ∼5% of the total CD4+ T cell response (Figure 6C). E, ORF6, hypothetical ORF10, and nsp1 are all small antigens (or potentially not expressed, in the case of ORF10) and were most likely predominantly unrecognized as a result. These results are somewhat unexpected, because data for other coronaviruses, from 27 different studies curated in the IEDB, reported that spike accounted for nearly two-thirds of reported CD4+ T cell reactivity (Table S3). N accounted for most of the remaining epitopes in the published literature, although human N-specific CD4+ T cell responses were not observed in one of the most comprehensive studies of human SARS-CoV-1 T cell responses (Li et al., 2008). Coronavirus M has not previously been described as a prominent target of CD4+ T cell responses (Table S3). In sum, these results, fully scanning the SARS2 orfeome, demonstrate a pattern of robust and diverse SARS-CoV-2-specific CD4+ T cell reactivity in convalescing COVID-19 cases that correlated largely with predicted viral protein abundance in infected cells.
Figure 6.
Protein Immunodominance of SARS-CoV-2-Specific CD4+ and CD8+ T Cells in COVID-19 Cases and Unexposed Donors
(A) SARS-CoV-2 genome organization and predicted viral protein abundance in infected cells.
(B) SARS-CoV-2 antigen-specific CD4+ T cells (AIM+, OX40+CD137+) quantified by stimulation index, using a peptide pool for each viral protein (with two exceptions, see Table S1). COVID-19 cases (top, in blue. n = 10) and unexposed donors (bottom, in white. n = 10). Data are expressed as geometric mean and geometric SD.
(C) Fraction of SARS-CoV-2 proteins recognized by CD4+ T cells in COVID-19 cases (top) and unexposed donors (bottom).
(D) SARS-CoV-2 antigen-specific CD4+ T cells (AIM+, OX40+CD137+) quantified by stimulation index, using a peptide pool for each viral protein (with two exceptions, see Table S1). COVID-19 cases (top, in red. n = 10) and unexposed donors (bottom, in gray. n = 10). Data are expressed as geometric mean and geometric SD.
(E) Fraction of SARS-CoV-2 proteins recognized by CD8+ T cells in COVID-19 cases (top) and unexposed donors (bottom).
See also Figures S6 and S7 and Table S6.
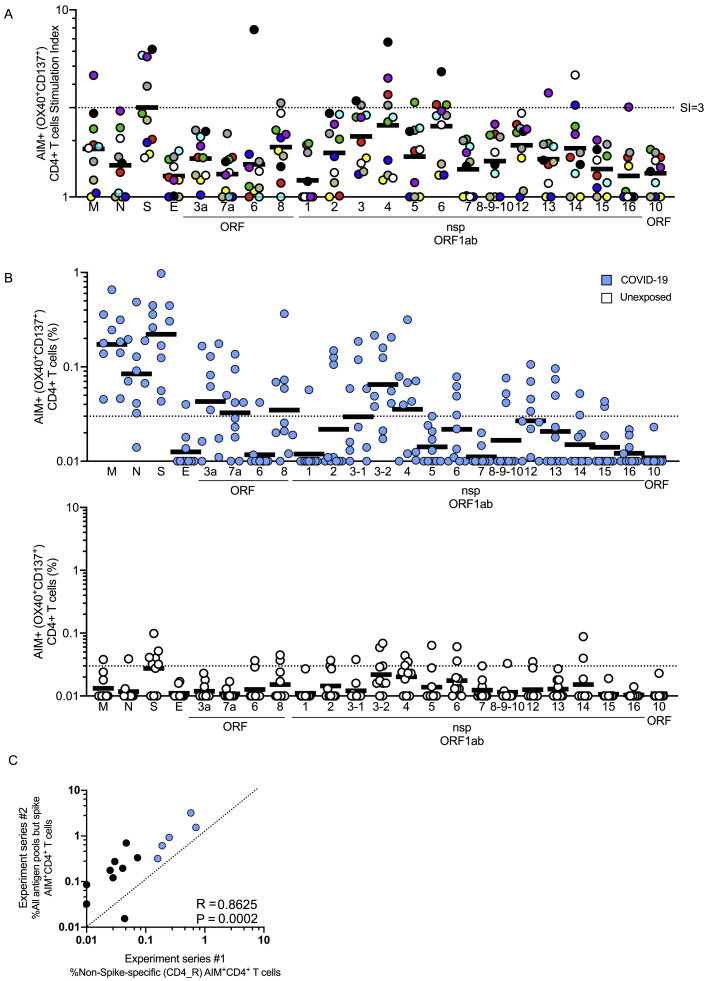
When examining the non-exposed donors, the pattern of CD4+ T cell targets changed. While S was still a relatively prominent target (23% of total, on average), there was no, or marginal, reactivity against SARS-CoV-2 N and M. Among donors with detectable CD4+ T cells, a shift in reactivity was observed toward SARS-CoV-2 nsp14 (25%), nsp4 (15%) and nsp6 (14%) (Figures 6B and 6C). SARS-CoV-2-reactive CD4+ T cells were detected in at least six different unexposed donors, demonstrating that the cross-reactivity is relatively widely distributed (Figure S6 A).
Figure S6.
Protein Immunodominance of SARS-CoV-2 Specific CD4+ T Cells in Recovered COVID-19 Patients and Unexposed Donors, Related to Figure 6
(A) The same data as Figure 6B, but with each unexposed donor color coded.
(B) The same experiment as Figure 6B, but with SARS-CoV-2−specific CD4+ T cells measured as percentage of AIM+ (OX40+CD137+) CD4+ T cells, after background subtraction. COVID-19 cases (top, in blue. n = 10) and unexposed donors (bottom, in white. n = 10).
(C) Correlation of SARS-CoV-2−specific CD4+ T cells detected using the epitope prediction approach (CD4_R MP) compared against the sum total of all antigen pools of overlapping peptides (excluding spike), run with samples from the same donors in two different experiment series. Dotted line indicates 1:1 concordance. Statistical comparison was performed using Spearman correlation.
Having scanned the full SARS-CoV-2 orfeome for CD4+ T cell reactivity in multiple donors, it was possible to assess whether the epitope prediction MP approach successfully enriched for SARS-CoV-2 epitopes targeted by human CD4+ T cells. When the total reactivity observed with the CD4_R MP was plotted versus the sum total of all antigen pools (excluding spike, given that spike predictions were not included in the CD4_R MP), a significant correlation was observed (p < 0.0002, Figure S6C). The single MP-R captured ∼50% (44% +/− range 28%–80%) of the non-spike response per COVID-19 donor, demonstrating the success of the prediction approach, which, as mentioned above, was devised to attempt to capture approximately 50% of the total response (Dhanda et al., 2018, Paul et al., 2015).
In the case of CD8+ T cell responses, the data in the literature from other coronaviruses (57 different studies curated in the IEDB; Table S3) reported spike accounting for 50% and N accounting for 36% of the defined epitopes. In a large study of human SARS-CoV-1 responses, spike was reported as essentially the only target of CD8+ T cell responses (Li et al., 2008), while in a study of MERS CD8+ T cells, responses were noted for spike, N and a pool of M/E peptides (Zhao et al., 2017). Few epitopes have been reported from other coronavirus antigens (Table S3). Here, we scanned the full SARS-CoV-2 orfeome for CD8+ T cell recognition. Our data indicate a somewhat different pattern of immunodominance for SARS-CoV-2 CD8+ T cell reactivity (Figures 6D and 6E), with spike protein accounting for ∼26% of the reactivity, and N accounting for ∼12%. Significant reactivity in COVID-19 recovered subjects was derived from other antigens, such as M (22%), nsp6 (15%), ORF8 (10%), and ORF3a (7%) (Figures 6D and 6E). In unexposed donors, SARS-CoV-2-reactive CD8+ T cells were detected in at least four different donors (Figure S7 ), with less clear targeting of specific SARS-CoV-2 proteins than was observed for CD4+ T cells, suggesting that coronavirus CD8+ T cell cross-reactivity exists but is less widespread than CD4+ T cell cross-reactivity.
Figure S7.
Protein Immunodominance of SARS-CoV-2-Specific CD8+ T Cells in Recovered COVID-19 Patients and Unexposed Donors, Related to Figure 6
(A) The same data as Figure 6D, but with each unexposed donor color coded.
(B) The same experiment as Figure 6D, but with SARS-CoV-2−specific CD8+ T cells measured as percentage of AIM+ (CD69+CD137+) CD8+ T cells, after background subtraction. COVID-19 cases (top, in red. n = 10) and unexposed donors (bottom, in gray. n = 10).
Discussion
There is a critical need for foundational knowledge about T cell responses to SARS-CoV-2. Here, we report functional validation of predicted epitopes when arranged in epitope MPs, utilizing PBMCs derived from convalescing COVID-19 cases. The experiments also used protein-specific peptide pools to determine which SARS-CoV-2 proteins are the predominant targets of human SARS-CoV-2-specific CD4+ and CD8+ T cells generated during COVID-19 disease. Importantly, we utilized the exact same series of experimental techniques with blood samples from healthy control donors (PBMCs collected in the 2015–2018 time frame), and substantial cross-reactive coronavirus T cell memory was observed.
Our results demonstrate that the epitope MPs are reagents well suited to analyze and detect SARS-CoV-2-specific T cell responses with limited sample material. We also developed and tested peptide pools corresponding to each of the 25 proteins encoded in the SARS-CoV-2 genome. Data from both the epitope MPs and protein peptide pool experiments can be interpreted in the context of previously reported T cell response immunodominance patterns observed for other coronaviruses, particularly the SARS and MERS viruses, which have been studied in humans, HLA-transgenic mice, wild-type mice, and other species. In the case of CD4+ T cell responses, data for other coronaviruses found that spike accounted for nearly two-thirds of reported CD4+ T cell reactivity, with N and M accounting for limited reactivity, and no reactivity in one large study of human SARS-CoV-1 responses (Li et al., 2008). Our SARS-CoV-2 data reveal that the pattern of immunodominance in COVID-19 is different. In particular, M, spike, and N proteins were clearly co-dominant, each recognized by 100% of COVID-19 cases studied here. Significant CD4+ T cell responses were also directed against nsp3, nsp4, ORF3s, ORF7a, nsp12, and ORF8. These data suggest that a candidate COVID-19 vaccine consisting only of SARS-CoV-2 spike would be capable of eliciting SARS-CoV-2-specific CD4+ T cell responses of similar representation to that of natural COVID-19 disease, but the data also indicate that there are many potential CD4+ T cell targets in SARS-CoV-2, and inclusion of additional SARS-CoV-2 structural antigens such as M and N would better mimic the natural SARS-CoV-2-specific CD4+ T cell response observed in mild to moderate COVID-19 disease.
Regarding SARS-CoV-2 CD8+ T cell responses, the pattern of immunodominance found here differed from the literature for other coronaviruses. However, stringent comparisons are not possible, as some earlier studies were not similarly comprehensive and did not utilize the same experimental strategy. The spike protein was a target of human SARS-CoV-2 CD8+ T cell responses, but it is not dominant. SARS-CoV-2 M was just as strongly recognized, and significant reactivity was noted for other antigens, mostly nsp6, ORF3a, and N, which comprised nearly 50% of the total CD8+ T cell response, on average. Thus, these data indicate that candidate COVID-19 vaccines endeavoring to elicit CD8+ T cell responses against the spike protein will be eliciting a relatively narrow CD8+ T cell response compared to the natural CD8+ T cell response observed in mild to moderate COVID-19 disease. An optimal vaccine CD8+ T cell response to SARS-CoV-2 might benefit from additional class I epitopes, such as the ones derived from the M, nsp6, ORF3a, and/or N.
There have been concerns regarding vaccine enhancement of disease by certain candidate COVID-19 vaccine approaches, via antibody-dependent enhancement (ADE) or development of a TH2 responses (Peeples, 2020). Herein, we saw predominant TH1 responses in convalescing COVID-19 cases, with little to no TH2 cytokines. Clearly more studies are required, but the data here appear to predominantly represent a classic TH1 response to SARS-CoV-2.
While it was important to identify antigen-specific T cell responses in COVID-19 cases, it is also of great interest to understand whether cross-reactive immunity exists between coronaviruses to any degree. A key step in developing that understanding is to examine antigen-specific CD4+ and CD8+ T cells in COVID-19 cases and in unexposed healthy controls, utilizing the exact same antigens and series of experimental techniques. CD4+ T cell responses were detected in 40%–60% of unexposed individuals. This may be reflective of some degree of cross-reactive, preexisting immunity to SARS-CoV-2 in some, but not all, individuals. Whether this immunity is relevant in influencing clinical outcomes is unknown—and cannot be known without T cell measurements before and after SARS-CoV-2 infection of individuals—but it is tempting to speculate that the cross-reactive CD4+ T cells may be of value in protective immunity, based on SARS mouse models (Zhao et al., 2016). Clear identification of the cross-reactive peptides, and their sequence homology relation to other coronaviruses, requires deconvolution of the positive peptide pools, which is not feasible with the cell numbers presently available, and time frame of the present study.
Regarding the value of cross-reactive T cells, influenza (flu) immunology in relationship to pandemics may be instructive. In the context of the 2009 H1N1 influenza pandemic, preexisting T cell immunity existed in the adult population, which focused on the more conserved internal influenza viral proteins (Greenbaum et al., 2009). The presence of cross-reactive T cells was found to correlate with less severe disease (Sridhar et al., 2013, Wilkinson et al., 2012). The frequent availability of cross-reactive memory T cell responses might have been one factor contributing to the lesser severity of the H1N1 flu pandemic (Hancock et al., 2009). Cross-reactive immunity to influenza strains has been modeled to be a critical influencer of susceptibility to newly emerging, potentially pandemic, influenza strains (Gostic et al., 2016). Given the severity of the ongoing COVID-19 pandemic, it has been modeled that any degree of cross-protective coronavirus immunity in the population could have a very substantial impact on the overall course of the pandemic, and the dynamics of the epidemiology for years to come (Kissler et al., 2020).
Limitations and Future Directions
Caveats of this study include the sample size and the focus on non-hospitalized COVID-19 cases. Sample size was limited by expediency. The focus on non-hospitalized cases of COVID-19 is a strength, in that these donors had uncomplicated disease of moderate duration, and thus it was encouraging that substantial CD4+ T cell and antibody responses were detected in all cases, and CD8+ T cell responses in the majority of cases. Complementing these data with MP T cell data from acute patients and patients with complicated disease course will also be of clear value, as will studies on the longevity of SARS-CoV-2 immunological memory. Additionally, lack of detailed information on common cold history or matched blood samples pre-exposure to SARS-CoV-2 prevents conclusions regarding the abundance of cross-reactive coronavirus T cells before exposure to SARS-CoV-2 and any potential protective efficacy of such cells. Finally, full epitope mapping in the future will add important detailed resolution of the human coronavirus-specific T cell responses.
In sum, we measured SARS-CoV-2-specific CD4+ and CD8+ T cells responses in COVID-19 cases. Using multiple experimental approaches, SARS-CoV-2-specific CD4+ T cell and antibody responses were observed in all COVID-19 cases, and CD8+ T cell responses were observed in most. Importantly, pre-existing SARS-CoV-2-cross-reactive T cell responses were observed in healthy donors, indicating some potential for pre-existing immunity in the human population. ORF mapping of T cell specificities revealed valuable targets for incorporation in candidate vaccine development and revealed distinct specificity patterns between COVID-19 cases and unexposed healthy controls.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| M5E2 (V500) [anti-CD14] | Becton Dickinson | 561391 (RRID:AB_10611856) |
| HIB19 (V500) [anti-CD19] | Becton Dickinson | 561121 (RRID:AB_10562391) |
| FN50 (BV605) [anti-CD4] | Becton Dickinson | 562989 (RRID:AB_2737935) |
| RPA-T8 (BV650) [anti-CD8] | BioLegend | 301042 (RRID:AB_2563505) |
| FN50 (PE-CF594) [anti-CD69] | Becton Dickinson | 562617 (RRID:AB_2737680) |
| Ber-ACT35 (PE-Cy7) [anti-OX40] | Biolegend | 350012 (RRID:AB_10901161)a |
| 4B4-1 (APC) [anti-CD137] | BioLegend | 309810 (RRID:AB_830672) |
| OKT3 (AF700) [anti-CD3] | Biolegend | 317340 (RRID:AB_2563408) |
| G043H7 (BV421) [anti-CD45RA] | BioLegend | 353207 (RRID:AB_10915137) |
| 4S.B3 (FITC) [anti-IFNγ] | Thermo Fisher Scientific | 11-7319-82 (RRID: AB_465415) |
| GB11 (PE) [anti-Granzyme B] | Thermo Fisher Scientific | 12-8899-41 (RRID: AB_1659718) |
| Mab11 (PeCy7) [anti-TNFα] | ebioscience | 25-7349-82 (RRID:AB_469686) |
| JES3-19F1 (APC) [anti-IL-10] | BioLegend | 506807 (RRID:AB_315457) |
| 3D12 (APC ef780) [anti-CCR7] | eBioscience | 47-1979-42 (RRID:AB_1518794) |
| B56 (FITC) [anti-KI67] | Becton Dickinson | 556026 (RRID:AB_396302) |
| SK3 (percp efluor710) [anti-CD4] | Invitrogen | 46-0047-42 (RRID:AB_1834401) |
| GB11 (af647) [anti-GzmB] | Biolegend | 515406 (RRID:AB_2566333) |
| MHM-88 (af700) [anti-IgM] | Biolegend | 314538 (RRID:AB_2566615) |
| O323 (APC cy7) [anti-CD27] | Biolegend | 302816 (RRID:AB_571977) |
| IA6-2 (PE) [anti-IgD] | Becton Dickinson | 555779 (RRID:AB_396114) |
| HCD56 (PE Dazzle) [anti-CD56] | Biolegend | 318348 (RRID:AB_2563564) |
| HIB19 (Cy5) [anti-CD19PE] | Biolegend | 302210 (RRID:AB_314240) |
| HIT2 (PECy7) [anti-CD38] | Invitrogen | 25-0389-42 (RRID:AB_1724057) |
| J252D4 (bv421) [anti-CXCR5] | Biolegend | 356920 (RRID:AB_2562303) |
| 63D3 (bv510) [anti-CD14] | Biolegend | 367123 (RRID:AB_2716228) |
| HI100 (bv570) [anti-CD45RA] | Biolegend | 304132 (RRID:AB_2563813) |
| G025H7 (bv605) [anti-CXCR3] | Biolegend | 353728 (RRID:AB_2563157) |
| 2H7 (bv650) [anti-CD20] | Biolegend | 302336 (RRID:AB_2563806) |
| G043H7 (bv711) [anti-CCR7] | Biolegend | 353228 (RRID:AB_2563865) |
| EH12.2H7 (bv786) [anti-PD-1] | Biolegend | 329930 (RRID:AB_2563443) |
| UCHT1 (buv395) [anti-CD3] | Becton Dickinson | 563546 (RRID:AB_2744387) |
| Live/dead (UV) [Zombie] | Biolegend | 423108 |
| 11A9 (buv496) [anti-CCR6] | Becton Dickinson | 612948 (RRID:AB_2833076) |
| 3G8 (buv737) [anti-CD16] | Becton Dickinson | 612786 (RRID:AB_2833077) |
| SK1 (buv805) [anti-CD8] | Becton Dickinson | 612889 (RRID:AB_2833078) |
| LEGENDplex 13-plex kit | Biolegend | 740809 |
| Biological Samples | ||
| Healthy donor blood samples | Carter BloodCare | http://www.carterbloodcare.org/ |
| Healthy donor blood samples | LJI Clinical Core | https://www.iedb.org/ |
| Convalescent donor blood samples | UC San Diego Health | http://www.health.ucsd.edu/ |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Synthetic peptides | Synthetic Biomolecules (aka A&A) | http://www.syntheticbiomolecules.com/ |
| SARS-CoV-2 Receptor Binding Domain (RBD) protein | Stadlbauer et al., 2020 | N/A |
| Critical Commercial Assays | ||
| CoronaCheck COVID-19 Rapid Antibody Test Kit | 20/20 BioResponse | https://coronachecktest.com/ |
| Deposited Data | ||
| Wuhan-Hu-1 RNA isolate | NCBI nuccore database | GenBank: MN_908947 |
| ORF10 protein | NCBI protein database | NCBI: YP_009725255.1 |
| Nucleocapsid phosphoprotein | NCBI protein database | NCBI: YP_009724397.2 |
| ORF8 protein | NCBI protein database | NCBI: YP_009724396.1 |
| ORF7a protein | NCBI protein database | NCBI: YP_009724395.1 |
| ORF6 protein | NCBI protein database | NCBI: YP_009724394.1 |
| membrane glycoprotein | NCBI protein database | NCBI: YP_009724393.1 |
| envelope protein | NCBI protein database | NCBI: YP_009724392.1 |
| ORF3a protein | NCBI protein database | NCBI: YP_009724391.1 |
| surface glycoprotein | NCBI protein database | NCBI: YP_009724390.1 |
| orf1ab polyprotein | NCBI protein database | NCBI: YP_009724389.1 |
| Software and Algorithms | ||
| IEDB | Vita et al., 2019 | https://www.iedb.org |
| IEDB-AR (analysis resource) | Dhanda et al., 2019 | http://tools.iedb.org |
| NetMHCpan EL 4.0 | Jurtz et al., 2017 | http://tools.iedb.org/mhci/ |
| IEDB | Vita et al., 2019 | https://www.iedb.org |
| Tepitool | Paul et al., 2016, Paul et al., 2015 | http://tools.iedb.org/tepitool/ |
| FlowJo 10 | FlowJo | https://www.flowjo.com/ |
| GraphPad Prism 8.4 | GraphPad | https://www.graphpad.com/ |
| LEGENDplex v8.0 | Biolegend | https://www.biolegend.com/ |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Alessandro Sette ([email protected]).
Materials Availability
Aliquots of synthesized sets of peptides utilized in this study will be made available upon request. There are restrictions to the availability of the peptide reagents due to cost and limited quantity.
Data and Code Availability
The published article includes all data generated or analyzed during this study, and summarized in the accompanying tables, figures and Supplemental materials.
Experimental Model and Subject Details
Human Subjects
Healthy Unexposed Donors
Samples from healthy adult donors were obtained by the La Jolla Institute for Immunology (LJI) Clinical Core or provided by a commercial vendor (Carter Blood Care) for prior, unrelated studies between early 2015 and early 2018. These samples were considered to be from unexposed controls, given that SARS-CoV-2 emerged as a novel pathogen in late 2019, more than one year after the collection of any of these samples. These donors were considered healthy in that they had no known history of any significant systemic diseases, including, but not limited to, autoimmune disease, diabetes, kidney or liver disease, congestive heart failure, malignancy, coagulopathy, hepatitis B or C, or HIV. An overview of the characteristics of these unexposed donors is provided in Table 1.
The LJI Institutional Review Board approved the collection of these samples (LJI; VD-112). At the time of enrollment in the initial studies, all individual donors provided informed consent that their samples could be used for future studies, including this study.
Convalescent COVID-19 Donors
The Institutional Review Boards of the University of California, San Diego (UCSD; 200236X) and La Jolla Institute (LJI; VD-214) approved blood draw protocols for convalescent donors. All human subjects were assessed for capacity using a standardized and approved assessment. Subjects deemed to have capacity voluntarily gave informed consent prior to being enrolled in the study. Individuals did not receive compensation for their participation.
Study inclusion criteria included subjects over the age of 18 years, regardless of disease severity, race, ethnicity, gender, pregnancy or nursing status, who were willing and able to provide informed consent, or with a legal guardian or representative willing and able to provide informed consent when the participant could not personally do so. Study exclusion criteria included lack of willingness or ability to provide informed consent, or lack of an appropriate legal guardian or representative to provide informed consent.
Blood from convalescent donors was obtained at a UC San Diego Health clinic. Blood was collected in acid citrate dextrose (ACD) tubes and stored at room temperature prior to processing for PBMC isolation and plasma collection. A separate serum separator tube (SST) was collected from each donor. Samples were de-identified prior to analysis. Other efforts to maintain the confidentiality of participants included referring to specimens and other records via an assigned, coded identification number.
Prior to enrollment in the study, donors were asked to provide proof of positive testing for SARS-CoV-2, and screened for clinical history and/or epidemiological risk factors consistent with the World Health Organization (WHO) or Centers for Disease Control and Prevention (CDC) case definitions of COVID-19 or Persons Under Investigation (PUI) (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/surveillance-and-case-definitions, https://www.cdc.gov/coronavirus/2019-nCoV/hcp/clinical-criteria.html). Per CDC and WHO guidance, clinical features consistent with COVID-19 included subjective or measured fever, signs or symptoms of lower respiratory tract illness (e.g., cough or dyspnea). Epidemiologic risk factors included close contact with a laboratory-confirmed case of SARS-CoV-2 within 14 days of symptom onset or a history of travel to an area with a high rate of COVID-19 cases within 14 days of symptom onset.
Disease severity was defined as mild, moderate, severe or critical based on a modified version of the WHO interim guidance, “Clinical management of severe acute respiratory infection when COVID-19 is suspected” (WHO Reference Number: WHO/2019-nCoV/clinical/2020.4). Mild disease was defined as an uncomplicated upper respiratory tract infection (URI) with potential non-specific symptoms (e.g., fatigue, fever, cough with or without sputum production, anorexia, malaise, myalgia, sore throat, dyspnea, nasal congestion, headache; rarely diarrhea, nausea and vomiting) that did not require hospitalization. Moderate disease was defined as the presence of lower respiratory tract disease or pneumonia without the need for supplemental oxygen, without signs of severe pneumonia, or a URI requiring hospitalization (including observation admission status). Severe disease was defined as severe lower respiratory tract infection or pneumonia with fever plus any one of the following: tachypnea (respiratory rate > 30 breaths per minute), respiratory distress, or oxygen saturation less than 93% on room air. Critical disease was defined as the need for ICU admission or the presence of acute respiratory distress syndrome (ARDS), sepsis, or septic shock, as defined in the WHO guidance document.
Convalescent donors were screened for symptoms prior to scheduling blood draws, and had to be symptom-free and approximately 3 weeks out from symptom onset at the time of the initial blood draw. Following enrollment, whole blood from convalescent donors was run on a colloidal-gold immunochromatographic ‘lateral flow’ assay to evaluate for prior exposure to SARS-CoV-2. This assay detects IgM or IgG antibodies directed against recombinant SARS-CoV-2 antigen labeled with a colloidal gold tracer (20/20 BioResponse CoronaCheck). Ninety percent of convalescent donors tested positive for IgM or IgG to SARS-CoV-2 by this assay (Table 1).
Convalescent donors were California residents, who were either referred to the study by a health care provider or self-referred. The majority (75%) of donors had a known sick contact with COVID-19 or suspected exposure to SARS-CoV-2 (Table 1). The most common symptoms reported were cough, fatigue, fever, anosmia, and dyspnea. Seventy percent of donors experienced mild illness. Donors were asked to self-report any known medical illnesses. Of note, 65% of these individuals had no known underlying medical illnesses.
Method Details
Peptide Pools
Epitope MegaPool (MP) design and preparation
SARS-CoV-2 virus-specific CD4 and CD8 peptides were synthesized as crude material (A&A, San Diego, CA), resuspended in DMSO, pooled and sequentially lyophilized as previously reported (Carrasco Pro et al., 2015). SARS-CoV-2 epitopes were predicted using the protein sequences derived from the SARS-CoV-2 reference (GenBank: MN908947) and IEDB analysis-resource as previously described (Dhanda et al., 2019, Grifoni et al., 2020). Specifically, CD4 SARS-CoV-2 epitope prediction was carried out using a previously described approach in Tepitool resource in IEDB (Paul et al., 2015, Paul et al., 2016), to select peptides with median consensus percentile ≤ 20, similar to what was previously described, but removing the resulting spike glycoprotein epitopes from this prediction (CD4-R (remainder) “Non-spike” MP, n = 221). This approach takes advantage of the extensive cross-reactivity and repertoire overlap between different HLA class II loci and allelic variants to predict promiscuous epitopes, capable of binding across the most common HLA class II prototypic specificities (Greenbaum et al., 2011, O’Sullivan et al., 1991, Sidney et al., 2010a, Sidney et al., 2010b, Southwood et al., 1998). The algorithm utilizes predictions for seven common HLA-DR alleles (DRB1∗03:01, DRB1∗07:01, DRB1∗15:01, DRB3∗01:01, DRB3∗02:02, DRB4∗01:01 and DRB5∗01:01) empirically determined to allow coverage of diverse populations and for different pathogens and antigen systems (Dhanda et al., 2018, Paul et al., 2015).
To investigate in-depth spike-specific CD4 T cells, 15-mer peptides (overlapping by 10 amino acids) spanning the entire antigen have been synthesized and pooled separately (CD-4 S (spike) MP, n = 253).
In the case of CD8 epitopes, since the overlap between different HLA class I allelic variants and loci is more limited to specific groups of alleles, or supertypes (Sidney et al., 2008), we targeted a set of the 12 most prominent HLA class I A and B alleles (A∗01:01, A∗02:01, A∗03:01, A∗11:01, A∗23:01, A∗24:02, B∗07:02, B∗08:01, B∗35:01, B∗40:01, B∗44:02, B∗44:03), which have been shown to allow broad coverage of the general population. CD8 SARS-CoV-2 epitope prediction was performed as previously reported, using NetMHC pan EL 4.0 algorithm (Jurtz et al., 2017) for the top 12 more frequent HLA alleles and selecting the top 1 percentile predicted epitope per HLA allele clustered with nested/overlap reduction (Grifoni et al., 2020). The 628 predicted CD8 epitopes were split in two CD8 MPs containing 314 peptides each (CD8-A and CD8-B). The CMV MP is a pool of previously reported class I and class II epitopes (Carrasco Pro et al., 2015).
Protein peptide pools
In the case of the protein pools, peptides of 15 amino acid length overlapping by 10 spanning each entire protein sequence were tested in a single MP (6-253 peptides per pool). Table S1 lists the number of peptides pooled for each of the viral proteins. Upon request we are prepared to make these MP available to the scientific community for use in a diverse set of investigations.
PBMC isolation
For all samples whole blood was collected in ACD tubes (COVID-19 donors) or heparin coated blood bag (healthy unexposed donors). Whole blood was then centrifuged for 15 min at 1850 rpm to separate the cellular fraction and plasma. The plasma was then carefully removed from the cell pellet and stored at −20C.
Peripheral blood mononuclear cells (PBMC) were isolated by density-gradient sedimentation using Ficoll-Paque (Lymphoprep, Nycomed Pharma, Oslo, Norway) as previously described (Weiskopf et al., 2013). Isolated PBMC were cryopreserved in cell recovery media containing 10% DMSO (GIBCO), supplemented with 10% heat inactivated fetal bovine serum, depending on the processing laboratory, (FBS; Hyclone Laboratories, Logan UT) and stored in liquid nitrogen until used in the assays.
SARS-CoV-2 RBD ELISA
SARS-CoV-2 Receptor Binding Domain (RBD) protein was obtained courtesy of Florian Krammer and Peter Kim (Stadlbauer et al., 2020). Corning 96-well half-area plates (ThermoFisher 3690) were coated with 1μg/mL SARS-CoV-2 RBD overnight at 4°C. ELISA protocol generally followed that of the Krammer lab, which previously demonstrated specificity (Stadlbauer et al., 2020). Plates were blocked the next day with 3% milk (Skim Milk Powder ThermoFisher LP0031 by weight/volume) in Phosphate Buffered Saline (PBS) containing 0.05% Tween-20 (ThermoScientific J260605-AP) for 2 hours at room temperature. Plasma was then added to the plates and incubated for 1.5 hours at room temperature. Prior to plasma addition to the plates, plasma was heat inactivated at 56°C for 30-60 minutes. Plasma was diluted in 1% milk in 0.05% PBS-Tween 20 starting at a 1:3 dilution and diluting each sample at by 1:3. Plates were then washed 5 times with 0.05% PBS-Tween 20. Secondary antibodies were diluted in 1% milk in 0.05% Tween-20 and incubated for 1 hour. For IgG, anti-human IgG peroxidase antibody produced in goat (Sigma A6029) was used at a 1:5000 dilution. For IgM, anti-human IgM peroxidase antibody produced in goat (Sigma A6907) was used at a 1:10,000 dilution. For IgA, anti-human IgA horseradish peroxidase antibody (Hybridoma Reagent Laboratory HP6123-HRP) was used at a 1:1,000 dilution. Plates were washed 5 times with 0.05% PBS-Tween 20. Plates were developed with TMB Substrate Kit (ThermoScientific 34021) for 15 minutes at room temperature. The reaction was stopped with 2M sulfuric acid. Plates were read on a Spectramax Plate Reader at 450 nm using SoftMax Pro, and ODs were background subtracted. A positive control standard was created by pooling plasma from six convalescing COVID-19 patients. Positive control standard was run on each plate and was used to calculate titers (relative units) for all samples using non-linear regression interpolations, done to quantify the amount of anti-RBD IgG, anti-RBD IgM, and anti-RBD IgA present in each specimen. Titers were plotted for each specimen and compared to COVID-19 negative specimens. As a second analytical approach, Area under the curve was also calculated for each specimen to compare COVID-19 to negative specimens, using a baseline of 0.05 for peak calculations.
OC43 and NL63 coronavirus RBD ELISA
An in-house ELISA at UNC was performed by coating with recombinant S RBD antigens (SARS-CoV-2, SARS-CoV, OC43-CoV and NL63-CoV) in TBS for 1 h at 37 C. After blocking, we added 1:20 diluted serum and incubated at 37°C for 1 h. Antigen-specific antibodies (Ig) were measured at 405 nm by using alkaline phosphatase conjugated goat anti-human IgG, IgA and IgM Abs and 4-Nitrophenyl phosphate.
Flow Cytometry
Direct ex vivo PBMC immune cell phenotyping
For the surface stain, 1x106 PBMCs were resuspended in 100 μl PBS with 2% FBS (FACS buffer) and stained with antibody cocktail for 1 hour at 4°C in the dark. Following surface staining, cells were washed twice with FACS buffer. Cells were then fixed/permeabilized for 40min at 4C in the dark using the eBioscience FoxP3 transcription factor buffer kit (ThermoFisher Scientific, Waltham, MA). Following fixation/permeabilization, cells were washed twice with 1x permeabilization buffer, resuspended in 100 μl permeabilization buffer and stained with intracellular/intranuclear antibodies for 1 hour at 4°C in the dark. Samples were washed twice with 1x permeabilization buffer following staining. After the final wash, cells were resuspended in 200μl FACS buffer. All samples were acquired on a BD FACSymphony cell sorter (BD Biosciences, San Diego, CA). A list of antibodies used in this panel can be found in Table S2.
T cell stimulations
For all flow cytometry assays of stimulated T cells, cryopreserved cells were thawed by diluting them in 10 mL complete RPMI 1640 with 5% human AB serum (Gemini Bioproducts) in the presence of benzonase [20ul/10mL]. All samples were acquired on a ZE5 Cell analyzer (Bio-rad laboratories), and analyzed with FlowJo software (Tree Star, San Carlos, CA).
Activation induced cell marker assay
Cells were cultured for 24 hours in the presence of SARS-CoV-2 specific MPs [1 μg/ml] or 10 μg/mL PHA in 96-wells U bottom plates at 1x106 PBMC per well. A stimulation with an equimolar amount of DMSO was performed as negative control, phytohemagglutinin (PHA, Roche, 1 μg/ml) and stimulation with a combined CD4 and CD8 cytomegalovirus MP (CMV, 1 μg/ml) were included as positive controls. Supernatants were harvested at 24 hours post-stimulation for multiplex detection of cytokines. Antibodies used in the AIM assay are listed in Table S4 . AIM assays shown in Figures 2 and 3 and AIM assays shown in Figure 6 had five COVID-19 donors in common and nine Unexposed donors. Full raw data is listed in Table S6 .
Intracellular cytokine staining assay
For the intracellular cytokine staining, PBMC were cultured in the presence of SARS-CoV-2 specific MPs [1 μg/ml] for 9 hours. Golgi-Plug containing brefeldin A (BD Biosciences, San Diego, CA) and monensin (Biolegend, San Diego, CA) were added 3 hours into the culture. Cells were then washed and surface stained for 30 minutes on ice, fixed with 1% of paraformaldehyde (Sigma-Aldrich, St. Louis, MO) and kept at 4°C overnight. Antibodies used in the ICS assay are listed in Table S5 . The gates applied for the identification of IFNγ, GzB, TNFα, or IL-10 production on the total population of CD8+ T cells were defined according to the cells cultured with DMSO for each individual.
Cytokine bead assays
Supernatants were collected from 24-hour stimulation cultures of the AIM assays and stored in 96 well plates at −20°C. Cytokines in cell culture supernatants of the same samples used for AIM were quantified using a human Th cytokine panel (13-plex) kit (LEGENDplex, Biolegend) according to the manufacturer’s instruction. Supernatants were mixed with beads coated with capture antibodies specific for IL-5, IL-13, IL-2, IL-6, IL-9, IL-10, IFNγ, TNFα, IL-17a, IL-17F, IL-4, IL-21 and IL-22 and incubated on a 96 well filter plate for 2 hours. Beads were washed and incubated with biotin-labeled detection antibodies for 1 hour, followed by a final incubation with streptavidin-PE. Beads were analyzed by flow cytometry using a FACS Canto cytometer. Analysis was performed using the LEGENDplex analysis software v8.0, which distinguishes between the 13 different analytes on basis of bead size and internal dye.
Identification of coronavirus epitopes and associated literature references
To identify coronavirus epitopes and associated references, the IEDB was searched (on April 16, 2020) utilizing the following queries. A first query was run to identify references associated with class I restricted CD8 epitopes, which utilized the criteria settings “Antigen”: Organism = Coronavirus (taxonomy ID 11118); “Assay”: Positive assays only; “Assay”: T cell assay; “MHC restriction” = MHC Class II; no parameters were defined for “Host” or “Disease.” This query identified 57 references, which are listed and displayed under the “References” tab on the results page.
A second query was run to identify references associated with class II restricted CD4 epitopes which utilized the criteria settings “Antigen”: Organism = Coronavirus (taxonomy ID 11118); “Assay”: Positive assays only; “Assay”: T cell assay; “MHC restriction” = MHC Class II; no parameters were defined for “Host” or “Disease.” This query identified 27 references, which are listed and displayed under the “References” tab on the results page.
A third query was run to specifically capture epitopes and map them back to the antigen of origin using the setting; “Antigen”: Organism = Coronavirus (taxonomy ID 11118); “Assay”: Positive assays only; “Assay”: T cell assay; no parameters were defined for “MHC restriction,” “Host” or “Disease.” Results were exported as csv files, and then examined in Excel to tabulate the number of CD4 and CD8 epitopes recognized in humans, mice, transgenic mice and other hosts associated with each respective antigen.
Quantification and Statistical Analysis
Data and statistical analyses were done in FlowJo 10 and GraphPad Prism 8.4, unless otherwise stated. The statistical details of the experiments are provided in the respective figure legends. Data plotted in linear scale were expressed as Mean + Standard Deviation (SD). Data plotted in logarithmic scales were expressed as Geometric Mean + Geometric Standard Deviation (SD). Correlation analyses were performed using Spearman, while Mann-Whitney or Wilcoxon tests were applied for unpaired or paired comparisons, respectively. Details pertaining to significance are also noted in the respective legends. T cell data have been calculated as background subtracted data or stimulation index. Background subtracted data were derived by subtracting the percentage of AIM+ cells after SARS-CoV-2 stimulation from the DMSO stimulation. Stimulation Index was calculated instead by dividing the percentage of AIM+ cells after SARS-CoV-2 stimulation with the percentage of AIM+ cells derived from DMSO stimulation. If the AIM+ cells percentage after DMSO stimulation was equal to 0, the minimum value across each cohort was used. When two stimuli were combined together, the percentage of AIM+ cells after SARS-CoV-2 stimulation was combined and either subtracted twice or divided by twice the value of the percentage of AIM+ cells derived from DMSO stimulation. Additional data analysis techniques are described in the STAR Methods sections above.
Acknowledgments
We would like to thank Cheryl Kim, director of the LJI flow cytometry core facility for outstanding expertise. We thank Prof. Peter Kim, Abigail Powell, PhD, and colleagues (Stanford) for RBD protein synthesized from Prof. Florian Krammer (Mt. Sinai) constructs. J.M. was supported by PhD student fellowships from the Departamento Administrativo de Ciencia, Tecnología e Innovación (COLCIENCIAS), and Pontificia Universidad Javeriana. This work was funded by the NIH NIAID under awards AI142742 (Cooperative Centers for Human Immunology) (S.C. and A.S.), National Institutes of Health contract Nr. 75N9301900065 (A.S. and D.W.), and U19 AI118626 (A.S. and B.P.). The BD FACSymphony purchase was partially funded by the Bill and Melinda Gates Foundation and LJI Institutional Funds (S.C. and A.S.). This work was additionally supported in part by the Johnathan and Mary Tu Foundation (D.M.S.), the NIAID under K08 award AI135078 (J.D.), and UCSD T32s AI007036 and AI007384 Infectious Diseases Division (S.I.R. and S.A.R.).
Author Contributions
Conceptualization, A.G., D.W., S.C., and A.S.; Investigation, A.G., D.W., J.M., C.R.M., J.M.D., D.M., L.P., R.S.J., A.S., and D.W.; Formal Analysis, A.G., D.W., C.R.M., J.M.D., J.M., and S.C.; Resources, S.I.R., S.A.R., D.M.S., A.F.C., F.K., S.C., and A.S.; Data Curation, J.A.G. and B.P.; Writing, S.C., A.S., A.G., and D.W.; Supervision, B.P., A.M.d.S., S.C., and A.S.; Project Administration, A.F.; Funding Acquisition, S.C., A.S., D.W., D.S., and J.D.
Declaration of Interests
The authors declare no competing interests.
Published: May 14, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.05.015.
Supplemental Information
References
- Alshukairi A.N., Zheng J., Zhao J., Nehdi A., Baharoon S.A., Layqah L., Bokhari A., Al Johani S.M., Samman N., Boudjelal M. High Prevalence of MERS-CoV Infection in Camel Workers in Saudi Arabia. MBio. 2018;9 doi: 10.1128/mBio.01985-18. e01985–e01918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Krammer F. SARS-CoV-2 Vaccines: Status Report. Immunity. 2020;52:583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S.F., Huang Y., Kaur K., Popova L.I., Ho I.Y., Pauli N.T., Henry Dunand C.J., Taylor W.M., Lim S., Huang M. Immune history profoundly affects broadly protective B cell responses to influenza. Sci. Transl. Med. 2015;7:316ra192. doi: 10.1126/scitranslmed.aad0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft T., Dillon M.B., da Silva Antunes R., Paul S., Peters B., Crotty S., Lindestam Arlehamn C.S., Sette A. Th1 versus Th2 T cell polarization by whole-cell and acellular childhood pertussis vaccines persists upon re-immunization in adolescence and adulthood. Cell. Immunol. 2016;304-305:35–43. doi: 10.1016/j.cellimm.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Møller R., Panis M., Sachs D., Albrecht R.A., tenOever B.R. SARS-CoV-2 launches a unique transcriptional signature from in vitro, ex vivo, and in vivo systems. bioRxiv. 2020 doi: 10.1101/2020.03.24.004655. [DOI] [Google Scholar]
- Callow K.A., Parry H.F., Sergeant M., Tyrrell D.A. The time course of the immune response to experimental coronavirus infection of man. Epidemiol. Infect. 1990;105:435–446. doi: 10.1017/s0950268800048019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X. COVID-19: immunopathology and its implications for therapy. Nat. Rev. Immunol. 2020;20:269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco Pro S., Sidney J., Paul S., Lindestam Arlehamn C., Weiskopf D., Peters B., Sette A. Automatic Generation of Validated Specific Epitope Sets. J. Immunol. Res. 2015;2015:763461. doi: 10.1155/2015/763461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe P.G., Perera R.A.P.M., Park W.B., Song K.H., Bang J.H., Kim E.S., Kim H.B., Ko L.W.R., Park S.W., Kim N.J. MERS-CoV Antibody Responses 1 Year after Symptom Onset, South Korea, 2015. Emerg. Infect. Dis. 2017;23:1079–1084. doi: 10.3201/eid2307.170310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. T Follicular Helper Cell Biology: A Decade of Discovery and Diseases. Immunity. 2019;50:1132–1148. doi: 10.1016/j.immuni.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Antunes R., Paul S., Sidney J., Weiskopf D., Dan J.M., Phillips E., Mallal S., Crotty S., Sette A., Lindestam Arlehamn C.S. Definition of Human Epitopes Recognized in Tetanus Toxoid and Development of an Assay Strategy to Detect Ex Vivo Tetanus CD4+ T Cell Responses. PLoS ONE. 2017;12:e0169086. doi: 10.1371/journal.pone.0169086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J.M., Lindestam Arlehamn C.S., Weiskopf D., da Silva Antunes R., Havenar-Daughton C., Reiss S.M., Brigger M., Bothwell M., Sette A., Crotty S. A Cytokine-Independent Approach To Identify Antigen-Specific Human Germinal Center T Follicular Helper Cells and Rare Antigen-Specific CD4+ T Cells in Blood. J. Immunol. 2016;197:983–993. doi: 10.4049/jimmunol.1600318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J.M., Havenar-Daughton C., Kendric K., Al-Kolla R., Kaushik K., Rosales S.L., Anderson E.L., LaRock C.N., Vijayanand P., Seumois G. Recurrent group A Streptococcus tonsillitis is an immunosusceptibility disease involving antibody deficiency and aberrant TFH cells. Sci. Transl. Med. 2019;11:eaau3776. doi: 10.1126/scitranslmed.aau3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanda S.K., Karosiene E., Edwards L., Grifoni A., Paul S., Andreatta M., Weiskopf D., Sidney J., Nielsen M., Peters B., Sette A. Predicting HLA CD4 Immunogenicity in Human Populations. Front. Immunol. 2018;9:1369. doi: 10.3389/fimmu.2018.01369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanda S.K., Mahajan S., Paul S., Yan Z., Kim H., Jespersen M.C., Jurtz V., Andreatta M., Greenbaum J.A., Marcatili P. IEDB-AR: immune epitope database-analysis resource in 2019. Nucleic Acids Res. 2019;47(W1):W502–W506. doi: 10.1093/nar/gkz452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M.E., Katsaounou P. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.04.009. Published online April 17, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorse G.J., Patel G.B., Vitale J.N., O’Connor T.Z. Prevalence of antibodies to four human coronaviruses is lower in nasal secretions than in serum. Clin. Vaccine Immunol. 2010;17:1875–1880. doi: 10.1128/CVI.00278-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostic K.M., Ambrose M., Worobey M., Lloyd-Smith J.O. Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science. 2016;354:722–726. doi: 10.1126/science.aag1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum J.A., Kotturi M.F., Kim Y., Oseroff C., Vaughan K., Salimi N., Vita R., Ponomarenko J., Scheuermann R.H., Sette A., Peters B. Pre-existing immunity against swine-origin H1N1 influenza viruses in the general human population. Proc. Natl. Acad. Sci. USA. 2009;106:20365–20370. doi: 10.1073/pnas.0911580106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum J., Sidney J., Chung J., Brander C., Peters B., Sette A. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics. 2011;63:325–335. doi: 10.1007/s00251-011-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Angelo M.A., Lopez B., O’Rourke P.H., Sidney J., Cerpas C., Balmaseda A., Silveira C.G.T., Maestri A., Costa P.R. Global Assessment of Dengue Virus-Specific CD4+ T Cell Responses in Dengue-Endemic Areas. Front. Immunol. 2017;8:1309. doi: 10.3389/fimmu.2017.01309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A Sequence Homology and Bioinformatic Approach Can Predict Candidate Targets for Immune Responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–680. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T., Fan Y., Chen M., Wu X., Zhang L., He T., Wang H., Wan J., Wang X., Lu Z. Cardiovascular Implications of Fatal Outcomes of Patients With Coronavirus Disease 2019 (COVID-19) JAMA Cardiol. 2020 doi: 10.1001/jamacardio.2020.1017. Published online March 27, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Guo Z., Duan C., Chen Z., Wang G., Lu Y., Li M., Lu J. Long-Term Persistence of IgG Antibodies in SARS-CoV Infected Healthcare Workers. medRxiv. 2020 doi: 10.1101/2020.02.12.20021386. [DOI] [Google Scholar]
- Hancock K., Veguilla V., Lu X., Zhong W., Butler E.N., Sun H., Liu F., Dong L., DeVos J.R., Gargiullo P.M. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N. Engl. J. Med. 2009;361:1945–1952. doi: 10.1056/NEJMoa0906453. [DOI] [PubMed] [Google Scholar]
- Havenar-Daughton C., Reiss S.M., Carnathan D.G., Wu J.E., Kendric K., Torrents de la Peña A., Kasturi S.P., Dan J.M., Bothwell M., Sanders R.W. Cytokine-Independent Detection of Antigen-Specific Germinal Center T Follicular Helper Cells in Immunized Nonhuman Primates Using a Live Cell Activation-Induced Marker Technique. J. Immunol. 2016;197:994–1002. doi: 10.4049/jimmunol.1600320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herati R.S., Muselman A., Vella L., Bengsch B., Parkhouse K., Del Alcazar D., Kotzin J., Doyle S.A., Tebas P., Hensley S.E. Successive annual influenza vaccination induces a recurrent oligoclonotypic memory response in circulating T follicular helper cells. Sci Immunol. 2017;2:eaag2152. doi: 10.1126/sciimmunol.aag2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz D., Seumois G., Gholami A.M., Greenbaum J.A., Lane J., White B., Broide D.H., Schulten V., Sidney J., Bakhru P. Lack of allergy to timothy grass pollen is not a passive phenomenon but associated with the allergen-specific modulation of immune reactivity. Clin. Exp. Allergy. 2016;46:705–719. doi: 10.1111/cea.12692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A.T., Garcia-Carreras B., Hitchings M.D.T., Yang B., Katzelnick L., Rattigan S.M., Borgert B., Moreno C., Solomon B.D., Rodriguez-Barraquer I. A systematic review of antibody mediated immunity to coronaviruses: antibody kinetics, correlates of protection, and association of antibody responses with severity of disease. medRxiv. 2020 doi: 10.1038/s41467-020-18450-4. 2020.2004.2014.20065771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irigoyen N., Firth A.E., Jones J.D., Chung B.Y., Siddell S.G., Brierley I. High-Resolution Analysis of Coronavirus Gene Expression by RNA Sequencing and Ribosome Profiling. PLoS Pathog. 2016;12:e1005473. doi: 10.1371/journal.ppat.1005473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurtz V., Paul S., Andreatta M., Marcatili P., Peters B., Nielsen M. NetMHCpan-4.0: Improved Peptide-MHC Class I Interaction Predictions Integrating Eluted Ligand and Peptide Binding Affinity Data. J. Immunol. 2017;199:3360–3368. doi: 10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissler S.M., Tedijanto C., Goldstein E., Grad Y.H., Lipsitch M. Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period. Science. 2020:eabb5793. doi: 10.1126/science.abb5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.K., Wu H., Yan H., Ma S., Wang L., Zhang M., Tang X., Temperton N.J., Weiss R.A., Brenchley J.M. T cell responses to whole SARS coronavirus in humans. J. Immunol. 2008;181:5490–5500. doi: 10.4049/jimmunol.181.8.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindestam Arlehamn C.S., McKinney D.M., Carpenter C., Paul S., Rozot V., Makgotlho E., Gregg Y., van Rooyen M., Ernst J.D., Hatherill M. A Quantitative Analysis of Complexity of Human Pathogen-Specific CD4 T Cell Responses in Healthy M. tuberculosis Infected South Africans. PLoS Pathog. 2016;12:e1005760. doi: 10.1371/journal.ppat.1005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wei Q., Lin Q., Fang J., Wang H., Kwok H., Tang H., Nishiura K., Peng J., Tan Z. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4:123158. doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morou A., Brunet-Ratnasingham E., Dubé M., Charlebois R., Mercier E., Darko S., Brassard N., Nganou-Makamdop K., Arumugam S., Gendron-Lepage G. Altered differentiation is central to HIV-specific CD4+ T cell dysfunction in progressive disease. Nat. Immunol. 2019;20:1059–1070. doi: 10.1038/s41590-019-0418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutaftsi M., Tscharke D.C., Vaughan K., Koelle D.M., Stern L., Calvo-Calle M., Ennis F., Terajima M., Sutter G., Crotty S. Uncovering the interplay between CD8, CD4 and antibody responses to complex pathogens. Future Microbiol. 2010;5:221–239. doi: 10.2217/fmb.09.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan D., Arrhenius T., Sidney J., Del Guercio M.F., Albertson M., Wall M., Oseroff C., Southwood S., Colón S.M., Gaeta F.C. On the interaction of promiscuous antigenic peptides with different DR alleles. Identification of common structural motifs. J. Immunol. 1991;147:2663–2669. [PubMed] [Google Scholar]
- Okba N.M.A., Raj V.S., Widjaja I., GeurtsvanKessel C.H., de Bruin E., Chandler F.D., Park W.B., Kim N.J., Farag E.A.B.A., Al-Hajri M. Sensitive and Specific Detection of Low-Level Antibody Responses in Mild Middle East Respiratory Syndrome Coronavirus Infections. Emerg. Infect. Dis. 2019;25:1868–1877. doi: 10.3201/eid2510.190051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S., Lindestam Arlehamn C.S., Scriba T.J., Dillon M.B., Oseroff C., Hinz D., McKinney D.M., Carrasco Pro S., Sidney J., Peters B., Sette A. Development and validation of a broad scheme for prediction of HLA class II restricted T cell epitopes. J. Immunol. Methods. 2015;422:28–34. doi: 10.1016/j.jim.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S., Sidney J., Sette A., Peters B. TepiTool: A Pipeline for Computational Prediction of T Cell Epitope Candidates. Curr. Protoc. Immunol. 2016;114:18.19.1–18.19.24. doi: 10.1002/cpim.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeples L. News Feature: Avoiding pitfalls in the pursuit of a COVID-19 vaccine. Proc. Natl. Acad. Sci. USA. 2020;117:8218–8221. doi: 10.1073/pnas.2005456117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss S., Baxter A.E., Cirelli K.M., Dan J.M., Morou A., Daigneault A., Brassard N., Silvestri G., Routy J.P., Havenar-Daughton C. Comparative analysis of activation induced marker (AIM) assays for sensitive identification of antigen-specific CD4 T cells. PLoS ONE. 2017;12:e0186998. doi: 10.1371/journal.pone.0186998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F., Lanzavecchia A., Araki K., Ahmed R. From vaccines to memory and back. Immunity. 2010;33:451–463. doi: 10.1016/j.immuni.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Moutaftsi M., Moyron-Quiroz J., McCausland M.M., Davies D.H., Johnston R.J., Peters B., Rafii-El-Idrissi Benhnia M., Hoffmann J., Su H.P. Selective CD4+ T cell help for antibody responses to a large viral pathogen: deterministic linkage of specificities. Immunity. 2008;28:847–858. doi: 10.1016/j.immuni.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severance E.G., Bossis I., Dickerson F.B., Stallings C.R., Origoni A.E., Sullens A., Yolken R.H., Viscidi R.P. Development of a nucleocapsid-based human coronavirus immunoassay and estimates of individuals exposed to coronavirus in a U.S. metropolitan population. Clin. Vaccine Immunol. 2008;15:1805–1810. doi: 10.1128/CVI.00124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J., Peters B., Frahm N., Brander C., Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008;9:1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J., Steen A., Moore C., Ngo S., Chung J., Peters B., Sette A. Divergent motifs but overlapping binding repertoires of six HLA-DQ molecules frequently expressed in the worldwide human population. J. Immunol. 2010;185:4189–4198. doi: 10.4049/jimmunol.1001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J., Steen A., Moore C., Ngo S., Chung J., Peters B., Sette A. Five HLA-DP molecules frequently expressed in the worldwide human population share a common HLA supertypic binding specificity. J. Immunol. 2010;184:2492–2503. doi: 10.4049/jimmunol.0903655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwood S., Sidney J., Kondo A., del Guercio M.F., Appella E., Hoffman S., Kubo R.T., Chesnut R.W., Grey H.M., Sette A. Several common HLA-DR types share largely overlapping peptide binding repertoires. J. Immunol. 1998;160:3363–3373. [PubMed] [Google Scholar]
- Sridhar S., Begom S., Bermingham A., Hoschler K., Adamson W., Carman W., Bean T., Barclay W., Deeks J.J., Lalvani A. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat. Med. 2013;19:1305–1312. doi: 10.1038/nm.3350. [DOI] [PubMed] [Google Scholar]
- Stadlbauer D., Amanat F., Chromikova V., Jiang K., Strohmeier S., Arunkumar G.A., Tan J., Bhavsar D., Capuano C., Kirkpatrick E. SARS-CoV-2 Seroconversion in Humans: A Detailed Protocol for a Serological Assay, Antigen Production, and Test Setup. Curr. Protoc. Microbiol. 2020;57:e100. doi: 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T., Kawakami C., Yamada S., Satoh R., Hohdatsu T. Antibody-dependent enhancement occurs upon re-infection with the identical serotype virus in feline infectious peritonitis virus infection. J. Vet. Med. Sci. 2008;70:1315–1321. doi: 10.1292/jvms.70.1315. [DOI] [PubMed] [Google Scholar]
- Thanh Le T., Andreadakis Z., Kumar A., Gómez Román R., Tollefsen S., Saville M., Mayhew S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19:305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- Tian Y., Grifoni A., Sette A., Weiskopf D. Human T Cell Response to Dengue Virus Infection. Front. Immunol. 2019;10:2125. doi: 10.3389/fimmu.2019.02125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., de Groot R.J., Harbour D.A., Dalderup M., Gruffydd-Jones T., Horzinek M.C., Spaan W.J. Early death after feline infectious peritonitis virus challenge due to recombinant vaccinia virus immunization. J. Virol. 1990;64:1407–1409. doi: 10.1128/jvi.64.3.1407-1409.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita R., Mahajan S., Overton J.A., Dhanda S.K., Martini S., Cantrell J.R., Wheeler D.K., Sette A., Peters B. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 2019;47(D1):D339–D343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D., Angelo M.A., de Azeredo E.L., Sidney J., Greenbaum J.A., Fernando A.N., Broadwater A., Kolla R.V., De Silva A.D., de Silva A.M. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc. Natl. Acad. Sci. USA. 2013;110:E2046–E2053. doi: 10.1073/pnas.1305227110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D., Cerpas C., Angelo M.A., Bangs D.J., Sidney J., Paul S., Peters B., Sanches F.P., Silvera C.G., Costa P.R. Human CD8+ T-Cell Responses Against the 4 Dengue Virus Serotypes Are Associated With Distinct Patterns of Protein Targets. J. Infect. Dis. 2015;212:1743–1751. doi: 10.1093/infdis/jiv289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson T.M., Li C.K.F., Chui C.S.C., Huang A.K.Y., Perkins M., Liebner J.C., Lambkin-Williams R., Gilbert A., Oxford J., Nicholas B. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat. Med. 2012;18:274–280. doi: 10.1038/nm.2612. [DOI] [PubMed] [Google Scholar]
- Wong C.K., Lam C.W., Wu A.K., Ip W.K., Lee N.L., Chan I.H., Lit L.C., Hui D.S., Chan M.H., Chung S.S., Sung J.J. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin. Exp. Immunol. 2004;136:95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases From the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Xie X., Muruato A., Lokugamage K.G., Narayanan K., Zhang X., Zou J., Liu J., Schindewolf C., Bopp N.E., Aguilar P.V. An Infectious cDNA Clone of SARS-CoV-2. Cell Host Microbe. 2020;27:841–848. doi: 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Van Rooijen N., Perlman S. Evasion by stealth: inefficient immune activation underlies poor T cell response and severe disease in SARS-CoV-infected mice. PLoS Pathog. 2009;5:e1000636. doi: 10.1371/journal.ppat.1000636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Perlman S. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J. Virol. 2010;84:9318–9325. doi: 10.1128/JVI.01049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Legge K., Perlman S. Age-related increases in PGD(2) expression impair respiratory DC migration, resulting in diminished T cell responses upon respiratory virus infection in mice. J. Clin. Invest. 2011;121:4921–4930. doi: 10.1172/JCI59777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Mangalam A.K., Channappanavar R., Fett C., Meyerholz D.K., Agnihothram S., Baric R.S., David C.S., Perlman S. Airway Memory CD4(+) T Cells Mediate Protective Immunity against Emerging Respiratory Coronaviruses. Immunity. 2016;44:1379–1391. doi: 10.1016/j.immuni.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Alshukairi A.N., Baharoon S.A., Ahmed W.A., Bokhari A.A., Nehdi A.M., Layqah L.A., Alghamdi M.G., Al Gethamy M.M., Dada A.M. Recovery from the Middle East respiratory syndrome is associated with antibody and T-cell responses. Sci. Immunol. 2017;2:eaan5393. doi: 10.1126/sciimmunol.aan5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all data generated or analyzed during this study, and summarized in the accompanying tables, figures and Supplemental materials.