Abstract
The study aims to assess the detection performance of a rapid primary screening technique for COVID‐19 that is purely based on the cough sound extracted from 2200 clinically validated samples using laboratory molecular testing (1100 COVID‐19 negative and 1100 COVID‐19 positive). Results and severity of samples based on quantitative RT‐PCR (qRT‐PCR), cycle threshold, and patient lymphocyte numbers were clinically labeled. Our suggested general methods consist of a tensor based on audio characteristics and deep‐artificial neural network classification with deep cough convolutional layers, based on the dilated temporal convolution neural network (DTCN). DTCN has approximately 76% accuracy, 73.12% in TCN, and 72.11% in CNN‐LSTM which have been trained at a learning rate of 0.2%, respectively. In our scenario, CNN‐LSTM can no longer be employed for COVID‐19 predictions, as they would generally offer questionable forecasts. In the previous stage, we discussed the exactness of the total cases of TCN, dilated TCN, and CNN‐LSTM models which were truly predicted. Our proposed technique to identify COVID‐19 can be considered as a robust and in‐demand technique to rapidly detect the infection. We believe it can considerably hinder the COVID‐19 pandemic worldwide.
Keywords: convolutional neural network, cough, COVID‐19, dilated, temporal
1. INTRODUCTION
COVID‐19 caused by severe acute respiratory syndrome coronavirus 2 (SARS‐COV‐2) is an infectious disease recognized in China in December 2019. It was announced by World Health Organization (WHO) as a global pandemic in March 2020. 1 It infects mucosa (mucous membrane) in the throat and causes respiratory tract infections along the path to the lungs. COVID‐19 spreads through the saliva droplets or from the nasal discharges of an infected person when they cough or sneeze. This infectious disease starts with mild fever, difficulty breathing, and tiredness. About 59% of the infected people have a dry cough. In a short time, the effects of the disease tend to affect millions of millions, causing fluctuations in countries' economies worldwide. Thus, causing a devastating impact on the welfare of people. 2 , 3 According to the statistics of World meter, there have been 394 426 790 COVID‐19 positive cases and 5 753 942 death cases recorded as of February 6, 2022.
In Reference [4], it is of utmost necessity to put forth a solution for the early diagnosis of the disease as we encounter devastating impacts due to COVID‐19 and witness a tragic loss of lives. The disease spreads and its effect can be annihilated through increasing the proficiency of pre‐screening and subsequent testing methodologies. This method would aid in the development of targeted solutions to solve this issue. From the beginning of this pandemic, artificial intelligence (AI) researchers worldwide have been trying to build an AI‐based testing and diagnosis tool for faster and quicker disease detection. 5 So far, COVID‐19 is being diagnosed either by RT‐PCR (reverse transcription polymerase chain) or using radiograph (X‐ray) and computerized tomography (CT) scan. This type of radiographical image‐based detection is considered a simple process suggested by medical centers for testing. Because they are more accurate than a swab test (RT‐PCR), these chest imaging techniques have been used to diagnose COVID‐19 in several cases.
Although RT‐PCR is the traditional method standardized for COVID‐19 diagnosis worldwide, its limitation makes it quite hard for prediction which is shown in Figure 2. The swab test is a time‐consuming manual process that does not provide an instant prediction and necessitates the careful handling of lab tools and kits. The supply of these kits would be difficult or insufficient in a country where billions of people reside, especially during crisis and pandemic situations. 6 Unlike any other laboratory or diagnostic method in healthcare centers, this technique is not free of errors. There is also a need for an expert technician to collect samples of the nasal mucosa. Because this is a manual process, the technician and patient must exercise caution to prevent disease transmission.
FIGURE 2.

Proposed system for identifying COVID‐19 using cough sound
Notably, several works have ensured that the RT‐PCR test has low sensitivity, about 30%–60%, with reduced accuracy in the COVID‐19 prediction. The detailed analysis of various studies proved that RT‐PCR produced contradicting results. 7 , 8 On the other hand, CT scan images have high recall values, but these models' specificity is relatively minor. These methods of testing, especially in early testing using RT‐PCR and CT scan, are thus expensive, time‐consuming, and may violate social distancing. Also, we are unaware when this pandemic will come to an end, so there comes the requirement for building alternate tools for diagnosis that overcome all the above‐discussed constraints and disadvantages deployed widely. 9 , 10
COVID‐19 is known to cause dangerous pulmonary disease symptoms, such as difficulty breathing and dry cough. 6 So, coughing is a common symptom of the disease, and thus the information extracted from cough samples could be used to predict the infectious disease in the body. These symptoms can build deep learning models to diagnose patients according to the trained recorded samples from healthy and sick individuals. 11 With technological advancements and progress in developing deep learning AI models, we can use these models as a screening tool to predict disease in individuals. 12 Without any doubt, we can accept that emerging tactics in AI would undoubtedly aid in developing a tool for the early diagnosis of COVID‐19 in both symptomatic and asymptomatic individuals, according to recent research. This is a beneficial solution because, even if the symptoms are not noticeable (in an asymptomatic person), the pathogens would still infect the host's body, causing changes identifiable by AI models. This is because AI models have the capabilities to learn and acknowledge acoustic features and distinguish cough samples of COVID‐positive from COVID‐negative or healthy ones. 12
MIT researchers have observed that asymptomatic people of the disease would vary from healthy people to coughing. Such variations are not figured out or clear to humans, but these features could be easily picked and identified by AI. 13 One of the most crucial challenges in this prediction is the right amount of dataset (both large and clean) to build a deep learning model that can make solid and accurate predictions on this pulmonary disease from the available cough recordings. Based on the result from these models, we could further approve the patient for tests like RT‐PCR or CT SCAN, and so on, already mentioned above. Moreover, cough‐based audio diagnosis is also a pre‐screening tool, cost‐effective, scalable, and potential one in our fight against COVID‐19 diagnosis. This article discusses the diagnostic methods of COVID‐19 from the cough sound recordings of individuals.
2. LITERATURE SURVEY
The utility of sound was recognized by researchers long back as a potential feature in the predictions of actions and health. It starts with external microphone recorders of digital stethoscopes for diagnosing the lungs and heartbeat sounds. These also allowed researchers to listen to and interpret various methodologies, such as magnetic resonance imaging (MRI) and sonography. It is easier to examine and interpret things. Current trends in identifying audio sounds and their interpretations have an impact on changing the scenario. Several studies have been conducted to determine whether it is possible to classify different coughs, such as wet/dry coughs and audio analysis of the depth of the cough sound in patients. Considering the pulmonary system, there are more types of diseases, such as lung infection, pneumothorax, bronchitis, asthma, and so on, which are identified by signal processing of the cough sounds.
In recent years, many works have been published on the detection of COVID using cough sounds, CT scans, and X‐rays. Several authors have attempted to build deep learning models which are a part of AI to diagnose the virus. 14 Yunlu et al. proposed a method to classify people infected with COVID‐19 on a large scale. This work can be used to identify different breathing patterns in individuals. In this article, the respiratory simulation (RS) model is introduced. It is used to fill up space in the copious training dataset. The results show that six distinct respiratory trends have precision, accuracy, recall, and F1 of about 94.5%, 94.4%, 95.1%, and 94.8%. The designed model has an enormous capacity to apply in large‐scale screening.
Korpas et al. 15 compared cough sounds recorded in a clinical setting with the results of spirometry tests. Spirometry is one of the most usual tests used to measure the patient's lung function and it measures the quantity of air inhaled by a person and exhaled. Korpas also performed signal processing on the cough sounds by visualizing the time domain waveforms and has concluded that the cough sounds contain unique information from the spirometry tests. The author has also shown that the spirometry results can be changed when they give some drugs administered to the patients, whereas analysis of the cough sound remains unchanged.
Santosh 16 proposed a model where signal and speech processing and image analysis methodologies are integrated. The author put forth speech processing techniques to predict tuberculosis in a patient, aiding doctors widely. This model recommends using the convolutional neural network used in making unbiased decisions. Abeyrate et al. 17 examined variation in coughs of pneumonia, coughs of asthma, the below figure, and bronchitis coughs. They reach 93% sensitivity and 54% specificity using various features like (i) statistics on time sequence and (ii) modeling formant‐frequency. They achieved the above features based on the parameters extraction from cough samples recorded as shown in Figure 1.
FIGURE 1.
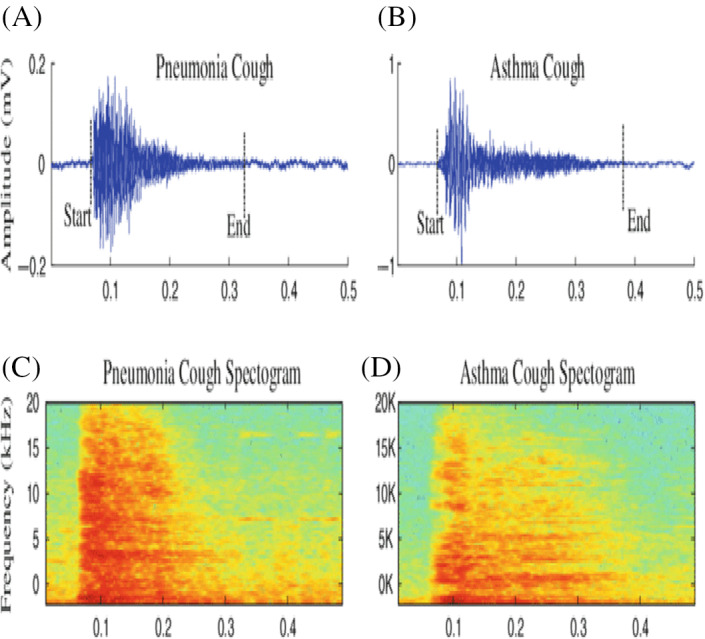
COVID‐19 cough samples
Swarkar et al. 18 had used various signal processing techniques to investigate the coughs sound. It includes analyzing properties like (i) energy spectrum and (ii) statistical waveform independent of time. The dataset of 536 samples could reach 54% recall and 92% specificity by splitting the dataset into dry coughs and wet coughs. Nallanthighal et al. 19 used CNN and RNN models or architectures to predict the results based on breath for breath as much as possible to predict asthma and general infections in the respiratory system.
Imran et al. 20 used convolutional neural network (CNN) architectures to perform the COVID‐19 to predict classifications based on the cough sounds recorded from various sources. The author has worked based on the pre‐trained model built from sound samples finely tuned on the dataset. In this article, the author considers transfer learning methods to perform at greater accuracy with an F1 score of 0.929. Abdelkader 21 has developed a complete‐fledged COVID‐19 analyzing method, and it can collect pieces of information, including cough samples, current temperature, and breath. Taking several parameters are input, it can classify them into various classes. The adaptative nature of the model enables it to be more suitable and can compute the early diagnosis of these respiratory illnesses. 22
Alphonse Pja improves the performance of diagnosing cough sound datasets by pre‐processing with a one‐dimensional (1D) convolution network. The author implemented an augmentation‐based mechanism in the model. Further, to extract the acoustic signal feature and in‐depth sound features that are given as input for one‐dimensional convolutional neural network (1D CNN), the data de‐noising auto encoder (DDAE) process is used rather than using the input as Mel‐frequency cepstral coefficient (MFCC) of cough samples. The accuracy and performance are also better than the previous models detecting the disease. Predictions are made from respiratory sounds using a 1D CNN classifier. Here, comprehensive classification including COVID‐19 cough sounds, asthma sounds, and normal healthy sounds are done with an accuracy of 90%. The author has discussed that there is a gain in accuracy of 4% when compared with MFCC.
In Reference [23], this study used a significant dataset for COVID‐19 based on infected people's breath and cough sounds. They include (i) COVID‐positive and COVID‐ negative patients' breath and cough analysis. (ii) COVID‐19‐positive and COVID‐19‐negative analysis with only cough sound. The task (1) achieved 80% accuracy with 220 on dealing with the combination of cough and breath, and task (2) achieved 82% accuracy with 29 users on dealing with a cough only. Recall function is slightly low at about (72%) because of not having specialized data to detect COVID‐19 cough properly. In this model, he used a support vector method (SVM) classifier to analyze the sound signals.
In Reference [24], Orlandic et al. created the dataset “COUGHVID” for analyzing the cough sound. In this dataset, details, such as gender, age, geographical locations, and COVID‐19 patients' health status are described with more than 20,000 cough sound recordings. We try to look at the self‐reported status variable which they have taken. It displays nutritional values with 25% of the recording sounds, COVID values with 25% of the recording sounds, symptomatic values with 35% of the recording sounds, and non‐reported status with 15% of the recording sounds. The percentage of COVID‐19‐positive symptoms in males is 65.5% and 34.5% in females, up from 7.5%, 15.5%, and 77% in males and females, respectively. Generally, features including cough, breath, and continuous speech are considered for identifying or analyzing COVID‐19, but they used cough sounds to detect the virus in this article.
To improve the quality of the image from high‐speed video endoscopy in shallow level light frames, the convolution model is being used. To classify the respiratory cysts through image datasets, detect tuberculosis (TB) through respiratory X‐ray images data and automatically make automatic nodules during endoscopy images. In developing the deep convolutional network, a chest X‐ray is becoming famous, and the result is shown in various applications. The machine‐learning model eased the method by convolution network with less quantity of pictures; otherwise, there is a need for a massive dataset to build the model. New studies for classification to detect the disease from limited X‐ray and CT scan data, 25 , 26 show improved results. However, with the increase in the number of datasets, we can achieve better accuracy.
They designed a model to diagnose and analyze COVID‐19 using the RNN model. Hassan demonstrated the impact of recurrent neural network (RNN) by using speech signal processing (SSP) to detect COVID disease, estimate the clear audible sounds of the patient's cough, breathing, and voice using LSTM for earlier screening, and diagnose the COVID‐19 virus. 27 The model finds poor quality in the speech test while comparing both cough and breath sound recordings. He performed an LSTM model, and it was obtained with less accuracy. They had proposed AI speech processing systems for COVID‐19 from cough recordings, and provide a personalized patient saliency map to longitudinally monitor patients in real‐time, noninvasively, and at essentially zero‐variable cost. 28 The proposed model achieves better accuracy with 100% accuracy.
They had explained a COVID‐19 patient cough and non‐COVID‐19 patient cough has been taken for the accuracy analysis. The AI4COVID‐19 app requires 2‐s cough video recording of the subject. The COVID‐19 problem has been solved by using a new multipronged intermediary centered risk‐averse AI structural design that lowers misdiagnosis. As a substitute, it gives a stability teletesting instrument deployable anytime, anywhere, by anyone, so clinical‐testing and treatment can be channeled to those who need it the most, thereby saving major life. 29
They had presented a machine‐learning‐based COVID‐19 cough classifier which can discriminate COVID‐19‐positive coughs from both COVID‐19‐negative and healthy coughs recorded on a smartphone. The result showed that the best performance was exhibited by the Resnet50 classifier, which was best able to discriminate between the COVID‐19‐positive and the healthy coughs with an area under the ROC curve (AUC) of 0.98. 30
For the automatic detection of COVID‐19 in raw audio files, the main objective of Alberto Tena et al. 31 is to design a freely available, quick, and efficient methodology. Toward diagnose of COVID‐19 using a supervised machine‐learning algorithm, this method is based on automated extraction of time–frequency cough features and selection of the more significant ones to be used. An accuracy of 90% was obtained.
In our work, we used a large dataset and trained it properly using the deep learning (DL) technique to achieve better results on the COVID‐19 cough sound dataset which is shown in Figures 2 and 3, respectively.
FIGURE 3.

Visualizing cough sound through waveform of a healthy individual and COVID‐19 affected person respectively (coughed three times)
3. PROPOSED MODEL
3.1. Temporal convolutional network
Temporal convolutional network (TCN) differs from CNN in terms of sequential modeling of tasks. It is quite a descriptive term for a family of architecture used. TCN provides more extended memory than recurrent architectures.
TCN employs a 1D fully connected convolutional network (FCN) architecture, with each hidden layer and input layer being the same size, and kernel size 1 being the length of zero paddings. This is added to make sure succeeding layers or stages have the same length as preceding layers. The TCN uses causal convolutions. It is convolution used exclusively in TCN, where output at a particular time (t) is convolved with elements from time (t) available in the preceding layer. TCN is a 1D FCN along with casual convolutions. Thus, its distinguishing features include (1) just like recurrent neural network, the TCN architecture takes up the length of any ordered series or sequence and maps it to a similar length output sequence, (2) the convolutions used in this architecture are causal so that there will be no data or information “leakage” from later to earlier, ensuring data protection. The TCN architecture must backpropagate to look for linear size available to the deep network and kernel or filter size as shown in Figure 4. So, keeping track of dependent parameters is relatively very challenging. Dilated convolutions could bring a solution to these problems.
FIGURE 4.

TCN architecture has 1 × 1 convolution along with a tubelet proposal
One‐dimensional convolutional network takes the input of a three‐dimensional tensor and outputs three‐dimensional tensor. Batch size, input length, input size are the input tensor of our TCN implementation model, and the batch size, input length, output size are the output tensor. Since every layer in a TCN has the same input and output length, only the third dimension of the input and output tensors differs among other parameters.
A single 1D convolutional layer with a batch size, input length, and input channels are the input tensor shape. As well as the batch size, input length, output channels are the outputs of a tensor. To analyze how a single layer converts its input into the output, we will have to look at one element of the batch (the same process occurs for every element in the batch when we consider the simplest case where input and output channels are both equal to 1. This is the case of 1‐D input and output tensors.
3.2. Dilated temporal convolution neural network
3.2.1. Dilated convolutions
When a 1D input sequence Y ∈ R n and filter in , the convolution operation of dilation of function U on element p is shown in Equation (1).
| (1) |
where ‘∙’represents element‐wise multiplication; l is the filter size or kernel size, df is the dilation parameter, or factor, specifies the direction of the previous by the dot product. Dilation is like becoming into a fixed or standard step in between two filter or kernel taps. The operation varies and becomes complex as the value of d changes. Especially, when dilation factor df is 1, a regular convolution is computed. When values of increase, the output is at the peak level with more significant range inputs. This aspect of TCN increases the width of the receptive region or field in any CNN. In our case, we can do this in two ways: (i) When higher values for l—filter size and df—dilation factor are provided, it appears to raise the width and expand the field of layers given by df for one layer. (ii) When dilated convolutions are generally used, the value of df (dilation factor) is raised concerning the depth of any network given by df is in order of where j is the levels which is shown in Figure 5.
FIGURE 5.

Dilated temporal convolution neural network
It is always necessary for dilated convolutional layers to get hold of large or wide receptive fields even if the parameters and number of layers are less. TCN block architecture, RES block architecture, and schematic representation of DTCN are clarified in Figure 6A–C, respectively.
FIGURE 6.

(A) TCN block architecture (B). RES block architecture (C). Schematic representation of DTCN
Dilated temporal convolution network structure depends on time sequences and their time‐based parameters is shown in Figure 6. It is an architecture with an N number of sequential blocks with different layers of layers in each block, forming a network. The input and output of dilated temporal convolution network architecture are sequentially associated with the time‐step. One‐dimensional convolution is performed in each layer in the blocks and the kernel (f) or filter of dilation 2n. Here n classifies the number of layers in each block. The operations of the dilation factor provided with high values ensure that the width of the receptive region or field is broad and comprehensive enough at the highest block. These fully connected convolution operations obtained the stabilization of the trained model as the complex computations are made concurrently or computed parallel.
Moreover, in a general temporal convolution network, residual blocks (ResBlock) are associated with layers of 1D convolutions with required filters and activation functions. There is a residual connection established between filters, with input and output layers. It can be noticed that the number of filters at the last block of this architecture is the same as the total number of feature matching at the output layer. Block two and block three are connected through a skip connection, as mentioned in the schematic diagram. This technique of dilation parameters and general TCN overcomes the limitations of vanishing gradient in the bottom layers of the model.
The recurrent networks are exclusively for sequential architectures dealing with time dependencies with a series of hidden layers. In the TCN paper, 32 the authors discussed a detailed comparison between the TCN and recurrent neural networks. The model's performance is computed for the various processes of a broad range, specifically for sequential networks like RNN.
Causal convolutional layers (dilated): Significant features of a TCN are dilated causal convolutional operations. “Causal” is termed as a filter or kernel at a particular time‐step (T1). These casual filters could visualize inputs that are less than time‐step (T1).
Residual blocks (ResBlock): In a particular Residual block, the above‐dilated casual convolutional layers are stacked, and the result or conclusion obtained from the top convolution block is summed up with the input layer to get the result of that block.
Aligning: Temporal convolution networks usually stack all the ResBlock to obtain more expansive receptive fields or regions which is shown in Figure 7. As a result, the TCN architecture and performance are comparable to that of a traditional recurrent network.
Calculation or computation of receptive field is needed to determine how many layers of residual blocks are required in the architecture.
FIGURE 7.

Residual blocks
3.3. Architectural elements
(a) Dilated: A temporal convolution network consisting of dilation parameters or factors df = 1, 2, and 4 with kernel or filter size l is 3 according to the figure below. The receptive blocks can include all the values in the input sequences mentioned. 33 , 34 , 35 , 36
(b) TCN Residual Blocks: In general, 1 × 1 convolution is added to inputs, and the output of residual layers has varied dimensions and features (dimension matching).
Residual functions and connections are represented in the figure. Black lines represent the 1 × 1 convolutions, and the blue lines represent the convolutional filters.
The number of previous time‐steps (history) is denoted as “j,” a dilated causal convolution layer.
For the 1st layer is given by, multiplied with dilation (m) where size is the size of the kernel or filter. It represents the preceding layer along with the location of the previous kernel subtracted with 1. The second layer, multiplied with dilation (m). Generally, multiplied with dilation (m). m is the total number of dilated convolutions from the initial or input layer. 37 , 38 In every residual block, the kernel or filter size and dilation factors are the same. So, the above equation can be reduced as , where m represents the mth residual block used. When the filter or size of kernel is constant, and if the dilation factor (df) of residual blocks is rising exponentially by k, dilation thus .
3.3.1. Convolutional neural network long short‐term memory
Convolutional neural network long short‐term memory (CNN‐LSTM) is an architecture, especially for sequence/series prediction problems. These sequential predictions result in spatial input, and LSTM is a suitable algorithm that is very similar to recurrent networks. In this, the model delivers the information of preceding layers to the next layer to keep track of past layers or stages as it is essential to make decisions suitable. Generally, for extracting features, the architecture makes use of CNN applied to input layers. The concept of CNN is combined with LSTM, that is, long short‐term memory, for manipulating sequential tasks. CNN‐LSTM architectures can be designed and implemented along with libraries like Keras. A CNN‐LSTM is constructed by connecting convolutional network layers and adding subsequent LSTM layers, as seen in the below diagram in Figure 8. This architecture is described in two models: (i) feature extraction using a convolution neural network architecture and (ii) LSTM layers for obtaining characteristics of time‐steps. The architecture diagram of the model includes the input layers, CNN layers (1 × 1 convolution) fully connected convolution layer, max‐pooling layers, LSTM models, and so on.
FIGURE 8.
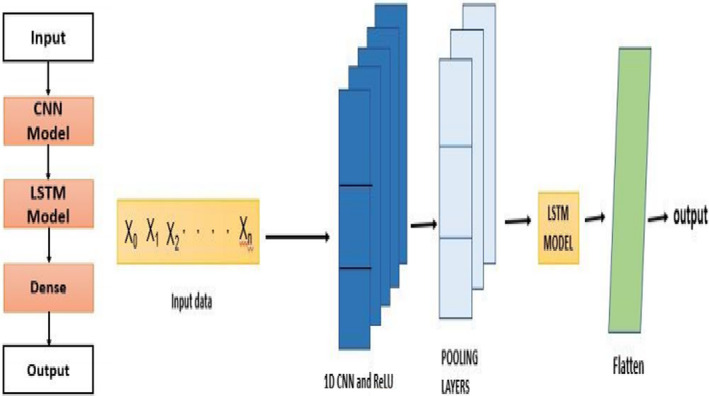
LSTM network
CNNs can synthesize relevant features and characteristics in a variety of ways; while the process of time sequencing (time series) can be done by LSTM architecture. The above two features are combined to design a CNN‐LSTM model like Recurrent Networks. LSTM is a time‐dependent architecture. 39 , 40 It also has time‐steps. Additionally, a component called “MEMORY” is available. Different functions used in LSTM are illustrated below.
(1) Forget gate —sigmoid activation layer in neural network. (2) Candidate layer —tanh activation layer. (3) Input Gate —sigmoid activation layer. (4) Output Gate “O”—sigmoid activation layer. (5) Hidden stage —vector to represent the state of hidden parameters. (6) Memory state —vector to represent the state to store values in each LSTM cell. Thus, the above all parameters except Candidate layer are all single‐layer or 1D neural networks with the sigmoid activation function, and only the Candidate layer belongs to the tanh activation function. 41 , 42 Each LSTM cell includes inputs and outputs with parameters mentioned below: Inputs include it “is the present input,” is the previously hidden stage, and is the previous memory state. Calculations are made concerning the time‐step (to). Once the inputs are given, the output of that cell includes (current hidden state) and (current state for memory). The weight vectors of gates are w and formed by performing dot operation for input vectors and past hidden stages and combining them. When activation functions including sigmoid and tanh are applied, they generate values in ranges of −1 to +1 in tanh and 0 and 1 for sigmoid. We will get the values of the four vectors as mentioned above in each cell of LSTM. The input is stored as a copy in the memory stage (M). To compute the current memory state, the memory state is multiplied by the forgetting gate (FG): The FG values are between 0 and 1. When or null, no previous stage is tracked, but when f g = 1, the content of the previous stage is passed. will be passed to the next step. Finally, calculations are made to reach the output. The output will be a filtered version of cell state . We apply tanh to , then we do multiplication with the output gate O, which is the currently hidden state are parameters given as inputs to the successive cell/next time‐step, and the same procedure repeats further.
3.4. Data collection
Collecting data is considered one of the essential processes in building a robust model that is trusted worldwide. The information is generally collected using sensors. Generally, different types of sensors are used to collect a variety of information. Gathering and organizing sound samples would need a microphone to record the cough samples from mobile phones or a web. 43 However, it becomes hard to gather and record a massive amount of information about this disease to build a robust model for better accuracy. We can recommend that people volunteer to record cough samples on available platforms through web browsers or developed apps. We can also collect the medical history of patients from hospitals, along with audio samples. It must be ensured that the cough sound recordings are done in a quiet environment to reduce unnecessary noise from the background. If the gathered data are accessible free (Open Source), researchers can use it to develop their models, trying to address or put forth solutions for the disease. For any data collection, proper approval is required from the person whose information is being recorded. To build a promising AI model for these pulmonary ailments, various features like body heat, mucus, etc., are collected and might be available in the corresponding metadata provided. The COVID‐19 pandemic and this devastating situation have proven the need for individuals to record and contribute to data collection to show various cough variations in the dataset. These measures will speed up the process by motivating patients and healthcare institutions, giving away their records to help researchers build the model which is shown in Figure 9. The work recommended in this article is to diagnose COVID‐19 using the COUGHVID dataset. The COUGHVID dataset is among the most extensive datasets consisting of cough samples available in open source. COUGHVID contains 20 000 recordings of cough sounds labeled as positive or unhealthy, symptomatic, or negative or healthy, and it contains the patient's clinical information in the metadata. 43 The dataset consists of an audio sample and necessary information about the patient like age, date and time of recording, gender, and existing symptoms—these features are used while building our model. We neglect the audio samples with a cough detection rate of less than 0.96, especially for negative coughs, giving us 4700 audio samples remaining. We considered 2200 voice recordings inclusive of both COVID‐19‐negative or healthy and COVID‐19‐positive or unhealthy cases. The database has two folders where these recordings are segregated into Positive and Negative cases, comprising 1100 recordings each. The dataset is now divided into two parts: training and testing, with 1000 samples for both positive and negative outcomes, a testing phase consisting of 100 samples for both positive and negative cases.
FIGURE 9.
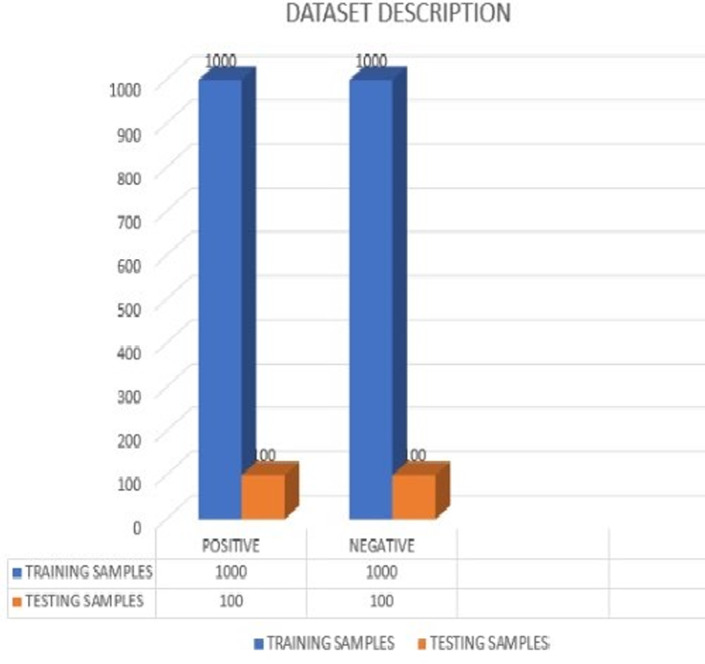
Data description of training and testing samples
4. RESULT AND DISCUSSION
This article deals with the adequate predictability of COVID‐19 from the recorded cough samples. The proposed normal TCN, Dilated TCN, and CNN‐LSTM architecture was trained with 2000 audio samples with positive and negative cases available in the COUGHVID database. The performance and result of the proposed model are tested for test audio samples. The TCN takes epochs to train the network. From the model accuracy and loss functions, it is evident that TCN, a simplified convolutional network architecture, performs better than recurrent networks like LSTM when predicted for a diverse dataset when working with a long memory as shown in Figure 10.
FIGURE 10.
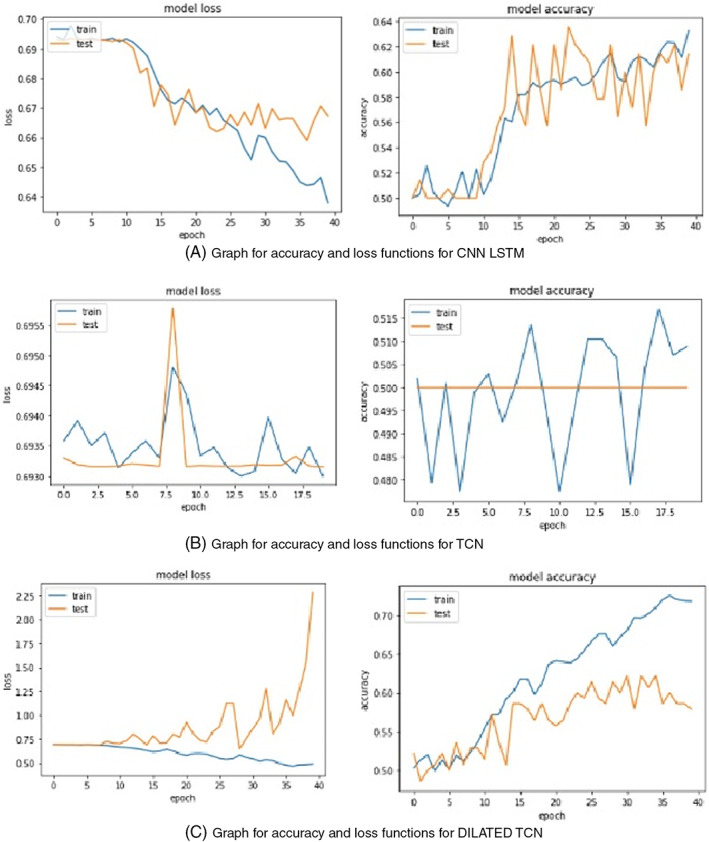
Training accuracy and loss for CNN‐LSTM, TCN, and DTCN
In DTCN, the accuracy manages to increase, and loss function decreases concerning epochs. Whereas, in CNN‐LSTM, 44 , 45 we could see that the accuracy varies in a zig–zag manner, and the loss rate is more significant than model accuracy, resulting in poor prediction. Even in the case of dilated TCN, the accuracy is comparatively lower than normal TCN. However, when we try to predict the accuracy of the models, various statistical metrics are accounted for to distinguish their features. One of the criteria for evaluating the success of any model is to analyze its accuracy. Thus, DTCN has an accuracy of about 76.7% and loss of about 48.6%, TCN has an accuracy of 73.12% and loss of 46.2%, and CNN‐LSTM with 72.11% accuracy and 50.3% loss. CNN‐LSTM has the low level of accuracy among other two models. In the previous step, we discussed the accuracy, which is genuinely predicted cases to the total cases for TCN, dilated TCN, and CNN‐LSTM models.
However, when the confusion matrix is plotted, 46 we can visualize the number of true‐positive, false‐positive, true‐negative, and false‐negative cases which is shown in Table 1. Moreover, other metrics can be calculated from confusion matrix parameters. Findings from the test set predicted by the proposed deep learning model allow us to find parameters, such as recall or sensitivity, F1‐score, precision. Precision is the ratio of predicted positive (true‐positive) cases to the total number of predicted positive cases.
TABLE 1.
Confusion matrix for test audio samples
| Actual class | Predicted class | ||
|---|---|---|---|
| Classification | COVID‐19 (Positive) | COVID‐19 (Negative) | |
| COVID‐19 (Positive) | 76 | 24 | |
| COVID‐19 (Negative) | 23 | 77 | |
Recall, also called sensitivity, is the ratio of exactly predicted positive (true‐positive) cases to the entire observations/cases in the actual class.
The F1‐score is known as the mean or average of recall/sensitivity and precision. We preferred to compute the confusion matrix and other parameters for the DTCN model as they show higher accuracy than TCN and CNN‐LSTM, they are calculated using the above formulas. From the calculations, the value of precision is 0.76, recall or sensitivity is 0.7676, and that of F1‐score is 0.76 which is shown in Figure 11.
FIGURE 11.

Performance metrics for different architectures
5. CONCLUSION
This article shows tremendous capabilities toward estimation and detection of COVID‐19 based on the recorded cough samples of individuals, with an overall accuracy of 76% for the TCN model. The cough sounds of both COVID‐19 patients and healthy individuals taken from the COUGHVID dataset were used for training and evaluating the performance of the architectures mentioned—features and characteristics being generated by the convolutional layer were used in the last layers of the network. In the model, maximum pooling of a 1D layer is being used. Accuracy of 76% in DTCN, 73.12% in TCN, and 72.11% in CNN‐LSTM, respectively were trained with a learning rate of 0.2. We successfully compared three different algorithms and concluded DTCN to be the best among the other two algorithms.
6. FUTURE WORKS
This work can be further extended to classify COVID‐19 cases by including various features and characteristics like patient breath, vocal sounds of an individual, spontaneous speech signals, and more accurate predictions. Several enhancements can be made to the architecture proposed to improve further. Increasing the dataset and providing clean audio samples, avoiding noise from the background, will provide higher performance. Extra efforts can be put forth to increase the accuracy further and understand various dependency parameters. This type of predictions would certainly help common people to access themselves using their cough samples when developed as an application. This is definitely a pre‐screening before approaching to hospitals and would escalate the efficiency of work and would aid healthcare department if deployed. It offers an diagnosis technique for useful to billions of people when reached out especially in populated countries to provide solution for the shortcomings in the limited number of tests and be available to everyone through mobile phones. This can be a reasonable contribution with artificial intelligence toward COVID‐19 helping individuals to take steps to protect themselves and for the safety of entire community. 7 , 32 , 47 , 48 , 49 , 50 , 51
AUTHOR CONTRIBUTIONS
Conceptualization: Gurram Sunitha and Rajesh Arunachalam. Formal Analysis and Investigation: Mohammed Abd‐Elnaby, Mahmoud Eid. Writing Original Draft Preparation: Gurram Sunitha, Rajesh Arunachalam, Mohammed Abd‐Elnaby, Mahmoud Eid and Ahmed Rashed. Writing‐Revise and Editing: Rajesh Arunachalam and Ahmed Rashed.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
ACKNOWLEDGMENT
The authors would like to acknowledge the support received from Taif University Researchers Supporting Project Number (TURSP‐2020/147), Taif University, Taif, Saudi Arabia.
Sunitha G, Arunachalam R, Abd‐Elnaby M, Eid MMA, Rashed ANZ. A comparative analysis of deep neural network architectures for the dynamic diagnosis of COVID‐19 based on acoustic cough features. Int J Imaging Syst Technol. 2022;32(5):1433‐1446. doi: 10.1002/ima.22749
[Corrections added on June 6, 2022, after first online publication: P.O. Box and postal code have been included in affiliations 2–4.]
Funding information Taif University, Grant/Award Number: TURSP‐2020/147; This Research was supported by Taif university Researchers Supporting Project Number (TURSP‐2020/147), Taif University, Taif, Saudia Arabia
DATA AVAILABILITY STATEMENT
Research data are not shared.
REFERENCES
- 1. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. De CMA, Lima O. Information about the new coronavirus disease (COVID‐19). Radiol Bras. 2020;53(2):131‐139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Struyf T, Deeks JJ, Dinnes J, et al. Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID‐19 disease. Cochrane Database Syst Rev. 2020;7(7):45‐52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liao J, Fan S, Chen J, et al. Epidemiological and clinical characteristics of COVID‐19 in adolescents and young adults. The Innovation. 2020;1(1):329‐337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ramesh G. Researcher at Bosch Centre for Data Science and AI at IIT Madras and Dr. SundeepTeki, a leader in AI and Neuroscience. https://www.kdnuggets.com/2020/12/covid-cough-ai-detecting-sounds.html
- 6. World Health Organization . WHO Coronavirus Disease (COVID‐19) Dashboard. WHO; 2020. [Google Scholar]
- 7. Ng MY, Lee EYP, Yang J, et al. Imaging profile of the COVID‐19 infection: radiologic findings and literature review. Radiol Cardiothoracic Imaging. 2020;2(1):e200034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang S, Kang B, Ma J, et al. A deep learning algorithm using CT images to screen for corona virus disease (COVID‐19). Eur Radiol. 2020;8:131‐138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lippi G, Plebani M. A six‐sigma approach for comparing diagnostic errors in healthcare—where does laboratory medicine stand? Ann Transl Med. 2018;6(10):35‐43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ai T, Yang Z, Hou H, et al. Correlation of chest CT and RT‐PCR testing for coronavirus disease 2019 (COVID‐19) in China: a report of 1014 cases. Radiology. 2020;296(2):32, 200642‐40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bewley T, Dubnov T, de Oliveira M. Signal analysis and classification of audio samples from individuals diagnosed with COVID‐19. Master of Science in Engineering Sciences (Mechanical Engineering), University of California, San Diego; 2020. https://escholarship.org/uc/item/29c2h9p9.
- 12. Chu J. Artificial intelligence model detects asymptomatic Covid‐19 infections through cellphone‐recorded coughs, MIT News Office. https://news.mit.edu/2020/covid-19-cough-cellphone-detection-102.
- 13. Mouawad P, Dubnov T, Dubnov S. Robust detection of COVID‐19 in cough sounds. SN Compt Sci. 2021;2:34. doi: 10.1007/s42979-020-00422-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang Y, Hu M, Li Q, et al. Abnormal respiratory patterns classifier may contribute to large‐scale screening of people infected with COVID‐19 in an accurate and unobtrusive manner. arXiv. 2002;45:5534. [Google Scholar]
- 15. Korpas J, Sadlonova J, Vrabec M. Analysis of the cough sound: an overview. Pulm Pharmacol. 1996;9(5):261‐268. [DOI] [PubMed] [Google Scholar]
- 16. Santosh KC. Speech processing in healthcare: can we integrate? In: Dey N, ed. Intelligent Speech Signal Processing. Elsevier; 2019:1‐4. [Google Scholar]
- 17. Abeyratne UR, Swarnkar V, Setyati A, Triasih R. Cough sound analysis can rapidly diagnose childhood pneumonia. Ann Biomed Eng. 2013;41(11):2448‐2462. [DOI] [PubMed] [Google Scholar]
- 18. Swarnkar V, Abeyratne UR, Chang AB, Amrulloh YA, Setyati A, Triasih R. Automatic identification of wet and dry cough in pediatric patients with respiratory diseases. Ann Biomed Eng. 2013;41(5):1016‐1028. [DOI] [PubMed] [Google Scholar]
- 19. Nallanthighal V, Härmä A, Strik H. Deep sensing of breathing signal during conversational speech. In: Proceedings of Interspeech 2019; 2019.
- 20. Imran A, Posokhova I, Qureshi HN, et al. AI4COVID‐19: Ai enabled preliminary diagnosis for covid‐19 from cough samples via an app. Inform Med Unlocked. 2020;9:456‐462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Belkacem AN, Ouhbi S, Lakas A, Benkhelifa E, Chen C. End‐to‐end AI‐based point‐of‐care diagnosis system for classifying respiratory illnesses and early detection of COVID‐19: a theoretical framework. Front Med. 2021;13:23‐35. doi: 10.3389/fmed.2021.585578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lella KK, Pja A. Automatic COVID‐19 disease diagnosis using 1D convolutional neural network and augmentation with human respiratory sound based on parameters: cough, breath, and voice. AIMS Public Health. 2021;8(2):240‐264. doi: 10.3934/publichealth.2021019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brown C, Chauhan J, Grammenos A, et al. Exploring automatic diagnosis of COVID‐19 from Crowdsourced Respiratory sound data. In: Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining; 2020.
- 24. Orlandic L, Teijeiro T, Atienza D. The COUGHVID crowdsourcing dataset: a corpus for the study of large scale cough analysis algorithms. Sci Data. 2009;22:11644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jain R, Gupta M, Taneja S, Hemanth DJ. Deep learning‐based detection and analysis of COVID‐19 on chest X‐ray images. Appl Intell. 2021;51:1690‐1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yazdani S, Minaee S, Kafieh R, et al. COVID CT‐net: predicting Covid‐19 from chest CT images using attentional convolutional network. arXiv. 2020;4:12‐18. [Google Scholar]
- 27. Hassan A, Shahin I, Alsabek MB. COVID‐19 detection system using recurrent neural networks. In: International Conference on Communications, Computing, Cybersecurity, and Informatics (CCCI). Sharjah, United Arab Emirates; 2020.
- 28. Laguarta J, Hueto F, Brian S. Subirana COVID‐19 artificial intelligence diagnosis using only cough recordings. IEEE Open J Eng Med Biol. 2020;1:275‐281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Imran A, Posokhova I, Qureshi HN, et al. AI4COVID‐19: AI enabled preliminary diagnosis for COVID‐19 from cough samples via an App. Inform Med Unlocked. 2020;34:1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pahar M, Klopper M, Warren R, Niesler T. COVID‐19 cough classification using machine learning and global smartphone recordings. Comput Biol Med. 2020;12:453‐460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tena A, Clarià F, Solsona F. Automated detection of COVID‐19 cough. Biomed Signal Process Control. 2022;71(1):103175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bai S, Kolter JZ, Koltun V. An empirical evaluation of generic convolutional and recurrent networks for sequence modeling. arXiv. 2018;13:4‐13. [Google Scholar]
- 33. Khalaf OI. Preface: Smart solutions in mathematical engineering and sciences theory. Math Eng Sci Aerosp. 2021;12(1):1‐4. [Google Scholar]
- 34. Khalaf OI, Sokiyna M, Alotaibi Y, Alsufyani A, Alghamdi S. Web attack detection using the input validation method: dpda theory. Comput Mater Contin. 2021;68(3):3167‐3184. [Google Scholar]
- 35. Suryanarayana G, Chandran K, Khalaf OI, Alotaibi Y, Alsufyani A, Alghamdi SA. Accurate magnetic resonance image super‐resolution using deep networks and Gaussian filtering in the stationary wavelet domain. IEEE Access. 2021;9:71406‐71417. doi: 10.1109/ACCESS.2021.3077611 [DOI] [Google Scholar]
- 36. Jebril IH, Hammad MA, Al Aboushi AR, Salahd MM, Khalaf OI. User satisfaction of electric‐vehicles about charging stations (home, outdoor, and workplace). Turk J Comput Math Educ. 2021;12(3):3589‐3593. doi: 10.17762/turcomat.v12i3.1637 [DOI] [Google Scholar]
- 37. Li G, Liu F, Sharma A. Research on the natural language recognition method based on cluster analysis using neural network. Math Probl Eng. 2021;21:9982305. doi: 10.1155/2021/9982305 [DOI] [Google Scholar]
- 38. Wisesa O, Andriansyah A, Khalaf OI. Prediction analysis for business to business (B2B) sales of telecommunication services using machine learning techniques. Majlesi J Elect Eng. 2020;14(4):145‐153. doi: 10.29252/mjee.14.4.145 [DOI] [Google Scholar]
- 39. Tavera CA, Ortiz JH, Khalaf OI, Saavedra DF, THH A. Wearable wireless body area networks for medical applications. Comput Math Methods Med. 2021;35:5574376. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40. Hoang AT, Nguyen XP, Khalaf OI, Tran TX, Chau MQ, et al. Thermodynamic simulation on the change in phase for carburizing process. Comput Mater Contin. 2021;68(1):1129‐1145. [Google Scholar]
- 41. Rajasoundaran S, Prabu AV, Subrahmanyam JBV, et al. Secure watchdog selection using intelligent key management in wireless sensor networks. Mater Today Proc. 2021;32:104‐113. doi: 10.1016/j.matpr.2020.12.1027 [DOI] [Google Scholar]
- 42. Dalal S, Khalaf OI. Prediction of occupation stress by implementing convolutional neural network techniques. J Inf Sci Eng. 2021;56:27‐42. doi: 10.4018/JCIT.20210701.oa3 [DOI] [Google Scholar]
- 43. Orlandic L, Teijeiro T, Atienza D. The COUGHVID crowdsourcing dataset: a corpus for the study of large‐scale cough analysis algorithms. arXiv. 2020;8:11644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sabeenian RS, Paramasivam ME, Anand R, Dinesh PM. Palm‐leaf manuscript character recognition and classification using convolutional neural networks. In: Vishwakarma HR, ed. Computing and Network Sustainability. Springer; 2019:397‐404. [Google Scholar]
- 45. Anand R, Sowmya V, Gopalakrishnan EA, Soman KP. Modified Vgg deep learning architecture for Covid‐19 classification using bio‐medical images. IOP Conf Ser: Mater Sci Eng. 2016;1084:12001. [Google Scholar]
- 46. Pranav JV, Anand R, Shanthi T, Manju K, Veni S, Nagarjun S. Detection and identification of COVID‐19 based on chest medical image by using convolutional neural networks. Int J Intell Netw. 2020;1:112‐118. [Google Scholar]
- 47. Worldometers . COVID‐19 Coronavirus pandemic, https://www.worldometers.info/coronavirus/?utm_campaign=homeAdvegas1
- 48. Gringel F. Temporal convolutional networks for sequence modeling; 2020. https://dida.do/blog/temporal-convolutional-networks-for-sequence-modeling
- 49. Lee C. [Tensorflow] Implementing temporal convolutional networks, understanding tensorflow part 3. https://medium.com/the-artificial-impostor/notes-understanding-tensorflow-part-3-7f6633fcc7c7
- 50. Shanthi T, Sabeenian RS, Anand R. Automatic diagnosis of skin diseases using convolution neural network. Microprocess Microsyst. 2020;76:103074. [Google Scholar]
- 51. Lu W, Li J, Li Y, Sun A, Wang J. A CNN‐LSTM‐based model to forecast stock prices. Complexity. 2020;2020:6622927. doi: 10.1155/2020/6622927 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Research data are not shared.


