Abstract
Cervical cancer is the most common cancer among women worldwide. The diagnosis and classification of cancer are extremely important, as it influences the optimal treatment and length of survival. The objective was to develop and validate a diagnosis system based on convolutional neural networks (CNN) that identifies cervical malignancies and provides diagnostic interpretability. A total of 8496 labeled histology images were extracted from 229 cervical specimens (cervical squamous cell carcinoma, SCC, n = 37; cervical adenocarcinoma, AC, n = 8; nonmalignant cervical tissues, n = 184). AlexNet, VGG-19, Xception, and ResNet-50 with five-fold cross-validation were constructed to distinguish cervical cancer images from nonmalignant images. The performance of CNNs was quantified in terms of accuracy, precision, recall, and the area under the receiver operating curve (AUC). Six pathologists were recruited to make a comparison with the performance of CNNs. Guided Backpropagation and Gradient-weighted Class Activation Mapping (Grad-CAM) were deployed to highlight the area of high malignant probability. The Xception model had excellent performance in identifying cervical SCC and AC in test sets. For cervical SCC, AUC was 0.98 (internal validation) and 0.974 (external validation). For cervical AC, AUC was 0.966 (internal validation) and 0.958 (external validation). The performance of CNNs falls between experienced and inexperienced pathologists. Grad-CAM and Guided Gard-CAM ensured diagnoses interpretability by highlighting morphological features of malignant changes. CNN is efficient for histological image classification tasks of distinguishing cervical malignancies from benign tissues and could highlight the specific areas of concern. All these findings suggest that CNNs could serve as a diagnostic tool to aid pathologic diagnosis.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10278-022-00722-8.
Keywords: Computer vision, Deep learning, Convolutional neural networks, Histopathological images, Cervical cancer
Introduction
Cervical cancer is the fourth most common cancer among women worldwide, responsible for approximately 570,000 new cases and 270,000 subsequent deaths annually [1]. The diagnosis of cervical cancer and classification of cancer types rely heavily on histopathology analysis by trained pathologists. Classification of cancer types is crucial as types behave differently in many ways [2]. Cervical squamous cell carcinoma (SCC) accounts for more than 75% of cervical cancer [3]. Results from randomized controlled trials have suggested that responses to specific treatments differ between cervical SCC and cervical adenocarcinoma (AC) [4]. However, the great gap between the dearth of pathologists and the increased demand for pathology has become a hindrance to efficient and accurate pathologic diagnosis. In developing countries, pathologists were overwhelmed by burdensome tasks and forced to produce professional decisions as quickly as possible [5]. As the result, conflicting or even incorrect results may occur due to carelessness or haste. A number of research showed that there is inter- or intra-observer variation in cervical specimen interpretation [6].
To facilitate the diagnosis process and improve the efficiency and accuracy of pathologic diagnosis, computer-aided diagnosis (CAD) has been applied to the diagnosis of several malignant cancers. A classification task that classifies endometrial nuclei and lesions by support vector machine (SVM) achieved an 85% accuracy and 72.05% sensitivity [7]. Although CAD has been proven to facilitate the diagnostic process, lacking feature extraction limits its usage. Traditional CADs based on decision tree or SVM are completely dependent on manual image features extraction, which is burdensome and prone to human mistakes. Deep learning, more specifically, convolutional neural network (CNN), achieves breakthroughs in image classification, namely improvement in model performance and exemption from handcrafted image features extraction [8]. CNN has already achieved expert-level performance in the domain of identification of some specific pathologies, including breast cancer, lung cancer, and colon disease. Ehteshami et al. have found that the performance of CNN was slightly higher than that of the pathologists for the binary classification task of distinguishing benign from malignant endometrial lesions [9]. Moreover, CNN could provide explainable evidence to both convince pathologists and assist clinicians in providing further medical care [10]. To date, much less attention has been given to distinguishing cervical malignancies by deep learning.
The objective of our study was to distinguish cervical cancer (SCC and AC) from benign cervical tissues by using pre-trained CNN models. In addition to histological image classification tasks, Gradient-weighted Class Activation Mapping (Grad-CAM) and Guided Grad-CAM were applied to provide diagnostic interpretability. All these attempts may help pathologists to improve diagnosis efficiency and accuracy and to interpret histological images effectively.
Method
Sample Collection and Preparation
Two hundred and twenty-nine (37 cervical SCC, 8 cervical AC, and 184 nonmalignant cervical tissues) histological sections of cervical specimens from January 2018 and December 2020 were acquired from the Department of Pathology, Xinhua hospital Chongming branch affiliated with Shanghai Jiaotong University. To ensure the generalizability of our methods, sections were divided into two groups: those obtained between January 2018 and December 2019 were used for model training and internal validation (24 cervical SCC, 6 cervical AC, and 131 nonmalignant cervical tissues); those acquired between January and December 2020 were used for external validation (13 cervical SCC, 2 cervical AC, and 53 nonmalignant cervical tissues). Specimens of cervical cancer were collected from patients with primary and untreated tumors. Specimens of nonmalignant cervical tissues were acquired from patients who underwent hysterectomy for non-cervical reasons (uterine fibroid, adenomyosis, and prolapse of the uterus). Specimens of nonmalignant cervical tissues collected from patients with abnormal TCT results or evidence of HPV infection or intraepithelial lesion were also excluded.
Preparation of H&E stained cervical tissue sections followed standard protocol. The first three steps, including fixation, dehydration, and paraffin embedding, were done manually by experienced pathologists. Dissolution of paraffin and H&E staining were done using Leica ASP300S and HistoCore SPECTRA ST Stainer (Leica Biosystems, Wetzlar, Germany). The clinical characteristics of patients (e.g., age, HPV infection, and menopausal status) were obtained.
Image Digitization and CNN Construction
Digital Imaging
To get as many images as possible for the further classification task, each section was digitalized into 26–87 (according to the size of the specimen) non-overlapping patches (2048 × 1536 pixels) at a × 20 objective lens using a Leica DM2500 optical microscope with a Leica DFC295 digital microscope color camera (Leica Microsystems, Wetzlar, Germany). All digitized images were screened by two professional pathologists to discard the low-quality images. Low quality was defined as follows: (1) image containing less than 50% information; (2) blurred image; (3) image with overlapped and/or folded tissue; and (4) image without reliable pathologic diagnosis. After screening, a total of 8496 histology images were saved in TIF format for further CNNs training and validation. The pathologic diagnosis of each image serves as the ground truth for the diagnostic classification.
Image Processing for Model Training
To meet the requirement of CNNs, each image was resized to 224 × 224 pixels to serve as an input image for the CNN. Random data augmentation was applied to each image of the training and interval validation to provide enhanced information. The items of augmentation included random geometric transformations, horizontal and vertical rotation, width and height change, and zoom. Also, we used the batch normalization (BN) method to normalize layer inputs.
CNN Algorithms for Classification
Four CNNs were used to figure out different histology image classification tasks, as they have been proven of great value in large-scale image classification. AlexNet was an early attempt to combine convolutions and max-pooling operations, characterized by the simplicity of architecture [11]. It was selected to make a comparison with the CNNs below. VGG19 consisted of a deep and homogeneous convolution structure. VGG19 was chosen as it was characterized by a very deep architecture [12]. ResNet-50 was characterized by a residual learning module, which enhances the depth of the model but lowers the complexity. It won 1st place on the ILSVRC 2015 classification task [13]. Xception was made up of separable convolution layers repeated multiple times with residual connections [14]. The basic configuration settings for CNNs were as follows: (1) loss function: binary cross-entropy; (2) optimizer: Adam; (3) batch-size: 32; (4) epoch: 50.
We deployed fivefold cross-validation to split the images collected between January 2018 and December 2019 into training and internal validation folds before model training, as it could minimize the selection bias. Three image classification tasks were set: (1) to distinguish cervical SCC from benign tissues; (2) to distinguish cervical AC from benign tissues; (3) to differentiate between cervical SCC and cervical AC.
Model Evaluation
Accuracy, precision, and recall were used to assess the performance of four CNNs. Besides, the area under the ROC curve (AUC) was calculated to evaluate the performance of CNNs. A ROC curve was generated by plotting sensitivity against (1 = specificity) at various threshold settings.
Visualization of the CNNs
To provide explainable evidence for pathologists to understand, Grad-CAM and Guided Grad-CAM have been used to highlight the regions of an image that were relevant to the given class. For a thorough explanation of Grad-CAM and guided backpropagation, in general, saw Zhou et al. [15] and Springenberg et al. [16], respectively. The saliency map, the oldest and the most frequently used explanation method for interpreting the predictions of CNNs, was used to make a comparison between Grad-CAM and Guided Grad-CAM.
Performance of Pathologists
To compare the performance of human experts with CNNs on image classification, six pathologists who were all currently active in clinical practice and had different levels of experience were recruited. Six pathologists were randomly divided into two groups. The first group, which included three pathologists (pathologist 1: resident pathologist with 4 years of resident experience; pathologist 2: resident pathologist with 5 years of practice experience; pathologist 3: attending pathologist with 8 years of practical experience), aimed to distinguish cervical SCC from benign tissues, and the second group (pathologist 4: resident pathologist who just finished a 3-year residency program; pathologist 5: associated chief pathologist with 8 years of practical experience; pathologist 6: associated chief pathologist specialized in gynecology pathology for 10 years) aimed to distinguish cervical AC from benign tissues. To mimic the procedure of CNN training and validation, experts’ classification results were obtained by a two-step method. In the first phase, the same sets of images used in model training were provided to four pathologists with labels on them. They were allowed to discuss with each other and consult with experts to solve any questions related to the diagnosis of cervical malignancy. After 1 week of training, every pathologist was asked to join the second phase. In the second phase, we asked every pathologist to assess the same sets of images used for external validation of CNNs and then divided images into two categories (for the first group: “benign” and “SCC”; for the second group: “benign” and “AC”). Each image can only be divided into one category. To mimic routine diagnostic pathology workflow, every pathologist assesses the images independently in 3 days. The pathologists’ classification results were converted into the operating points.
Implementation Environment
All the classification and visualization tasks were performed on a PC with a Windows 10 (64-bit) operation system, a single CPU (AMD Ryzen7 5900x), and a GPU (NVIDIA GeForce RTX 3090, 24 GB), 64 GB of system memory. The source code was written in Python 3.7.0. Other libraries used to process and classify histology image included Nvidia-smi 512.15; CUDA 11.6; Keras 2.7.0; numpy 1.20.3; matplotlib 3.5.0; OpenCV 4.5.4.60; seaborn 0.11.2; TensorFlow 2.7.0; scikit-learn 1.0.1.
Results
Patient Characteristics
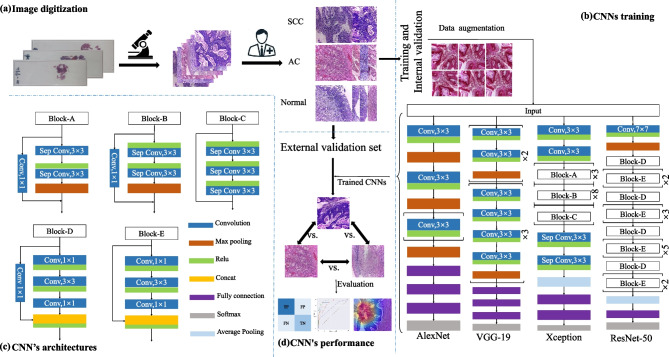
The characteristics of patients were shown in Tables S1, S2, and S3. In the benign group, the mean age of patients was 55.1 years. 100 (54.3%) patients were premenopausal and the major reason for their hysterectomy was fibrosis. All patients were diagnosed with chronic cervicitis. In the cervical SCC group, the mean age was 56.4 years, and 11 patients were premenopausal. According to FIGO classification, 28 patients (75.6%) were at stage I, 7 patients were at stage II, and 2 patients were at stage III. In the cervical AC group, the mean age was 48.3 years, and 5 patients were premenopausal. The stage of cervical AC was FIGO stage I in 7 patients (87.5%) and stage II in 1 patient (13.5%). The method’s flowchart is depicted in Fig. 1.
Fig. 1.
Study workflow. a Sample preparation and Image digitization; b model training and internal validation; c the architecture of AlexNet, VGG19, ResNet-50, and Xception; d CNN’s evaluation on external validation set
CNN Classified Cervical Cancer from Benign Tissues
The results of three binary classification tasks are listed in Table 1. All CNNs successfully distinguished cervical SCC from benign cervical tissues, achieving their respective AUC in the internal validation set as follows (AlexNet: 0.889 ± 0.001; ResNet: 0.955 ± 0.003; VGG19: 0.943 ± 0.002; Xception: 0.97 ± 0.003). Xception had the best performance in the internal validation set, followed by ResNet-50, VGG-19, and AlexNet. AlexNet got a worse performance than the remaining three CNNs in the internal validation set. Comparison of accuracy, precision, recall, loss, and AUC curves of four CNNs are shown in Fig. S1. Accuracy, precision, and AUC curves rose over time while the loss curve fell over time, and all eventually flattened out, indicating that there was no risk of overfitting.
Table 1.
The performance of CNNs for the internal and external validation set
| Classification tasks | CNN | Internal validation | External validation | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Accuracy | Precision | Recall | Auc | Accuracy | Precision | Recall | Auc | ||
| SCC vs. normal | AlexNet | 0.799 ± 0.01 | 0.814 ± 0.035 | 0.841 ± 0.058 | 0.889 ± 0.001 | 0.823 | 0.917 | 0.805 | 0.832 |
| Xception | 0.937 ± 0.008 | 0.941 ± 0.007 | 0.948 ± 0.018 | 0.98 ± 0.003 | 0.976 | 0.982 | 0.981 | 0.974 | |
| ResNet-50 | 0.878 ± 0.008 | 0.899 ± 0.019 | 0.885 ± 0.014 | 0.955 ± 0.003 | 0.938 | 0.966 | 0.939 | 0.938 | |
| VGG-19 | 0.866 ± 0.005 | 0.884 ± 0.007 | 0.880 ± 0.011 | 0.943 ± 0.002 | 0.932 | 0.937 | 0.961 | 0.918 | |
| AC vs. normal | AlexNet | 0.794 ± 0.015 | 0.828 ± 0.026 | 0.905 ± 0.049 | 0.845 ± 0.01 | 0.858 | 0.876 | 0.95 | 0.749 |
| Xception | 0.945 ± 0.004 | 0.962 ± 0.009 | 0.962 ± 0.009 | 0.966 ± 0.002 | 0.973 | 0.979 | 0.986 | 0.958 | |
| ResNet-50 | 0.893 ± 0.008 | 0.909 ± 0.011 | 0.947 ± 0.011 | 0.95 ± 0.006 | 0.951 | 0.969 | 0.982 | 0.915 | |
| VGG-19 | 0.887 ± 0.004 | 0.901 ± 0.006 | 0.948 ± 0.009 | 0.946 ± 0.003 | 0.938 | 0.961 | 0.978 | 0.889 | |
| SCC vs. AC | AlexNet | 0.697 ± 0.021 | 0.755 ± 0.027 | 0.785 ± 0.088 | 0.732 ± 0.019 | 0.779 | 0.802 | 0.934 | 0.630 |
| Xception | 0.925 ± 0.019 | 0.935 ± 0.018 | 0.95 ± 0.034 | 0.95 ± 0.014 | 0.960 | 0.969 | 0.976 | 0.944 | |
| ResNet-50 | 0.872 ± 0.014 | 0.890 ± 0.021 | 0.913 ± 0.015 | 0.943 ± 0.01 | 0.911 | 0.935 | 0.946 | 0.878 | |
| VGG-19 | 0.855 ± 0.011 | 0.849 ± 0.011 | 0.941 ± 0.011 | 0.931 ± 0.11 | 0.922 | 0.963 | 0.93 | 0.914 | |
AC adenocarcinoma, CNN convolutional neural networks, SCC squamous cell carcinoma
For the task of distinguishing cervical AC from benign tissues, Xception also achieved the best performance among four CNNs, with an accuracy of 0.945 ± 0.004, precision of 0.962 ± 0.009, recall of 0.962 ± 0.009, and AUC of 0.966 ± 0.002. The AUC of Xception was ~ 0.5% lower in distinguishing cervical AC than cervical SCC from benign tissues, while the AUC of AlexNet was ~ 4.5% lower when performing said tasks. A comparison of accuracy, precision, recall, loss, and AUC curves of four CNNs is shown in Fig. S2. The results suggested no evidence of overfitting.
As shown in Table 1, Xception also performed best in the binary classification task of distinguishing cervical SCC from cervical AC, with an accuracy of 0.943 ± 0.81%. A comparison of accuracy, precision, recall, loss, and AUC curves of four CNNs is shown in Fig. S3.
All CNNs were tested with the external validation set, with the resulting accuracy, precision, recall, and AUC values shown in Table 1 and Figs. S4, S5, and S6. Again, Xception achieved the best performance among the four CNNs, with an AUC of 0.974 for distinguishing SCC from benign tissues, 0.958 for distinguishing AC from benign tissues, and 0.928 for distinguishing SCC from AC. The ROC curve of AlexNet, VGG-19, ResNet-50, and Xception on external validation set along with the operating points of the pathologists is shown in Fig. 2. In task one, pathologist 3 had slightly better performance than that of CNNs. In task two, two pathologists (5 and 6) had slightly better performance than that of CNNs. However, diagnostic discrepancy among pathologists was significant. Pathologist 5 achieved a similar sensitivity, accuracy, and precision to pathologist 6, both had better performance than pathologist 4.
Fig. 2.
Receiver-operating characteristic (ROC) curves of four CNNs and the operating points of the pathologists. a ROC curves for distinguishing cervical SCC from benign tissues; b ROC curves for distinguishing cervical AC from benign tissues; c ROC curves for distinguishing cervical SCC from cervical AC
CNN’s Visualization
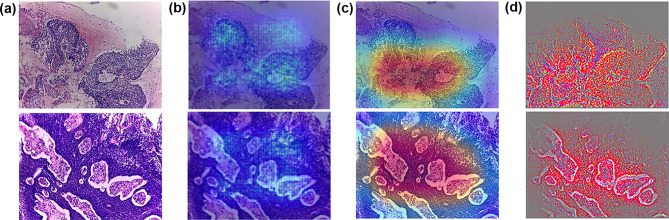
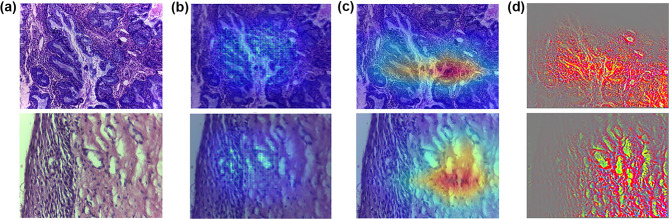
Figures 3 and 4 display the H&E images and the results of the visualization techniques of Xception. Figure 3(a) presents two histology images of cervical SCC. The H&E images showed solid sheets separated by an intervening desmoplastic or inflammatory stroma and network pattern infiltrating. Moreover, nuclei of squamous epithelial cells exhibited large and hyperchromatic with coarse chromatin. The saliency map, Grad-CAMs, and Guided Grad-CAMs all captured infiltrating and solid sheets that can distinguish SCC from benign tissues (Fig. 3b, c, and d). Compared with the saliency map, Grad-CAM and Guided Grad-CAM highlighted the malignant regions more accurately and displayed these regions in gradient mode with high resolution. The H&E images in Fig. 4(a) displayed two examples of cervical AC. Clear contours of individual glands appeared to be almost lost. The phenomenon of “back-to-back” (B2B) glands with little intervening stroma and diffusely infiltrative glands with associated extensive desmoplastic response were also identified. The results showed that the saliency map, Grad-CAMs, and Guided Grad-CAMs all highlighted the malignant changes of cervical AC, but later two methods had higher resolution (Fig. 4b, c, and d). Figure 5 displays two benign images that were misclassified as malignant by Xception. The Saliency map, Grad-CAM, and Guided Grad-CAM showed visually why Xception misclassified these two images. In the upper images of Fig. 5(b) and (c), Grad-CAM and Guided Grad-CAM highlighted the endocervical glands located under the squamous epithelium. In the lower images of Fig. 5(b) and (c), Xception was unable to identify the squamous epithelium region.
Fig. 3.
H&E, Grad-CAM, and Guided Grad-CAM of Xception model of cervical SCC. a H&E; b Grad-CAM; c Guided Grad-CAM
Fig. 4.
H&E, Grad-CAM, and Guided Grad-CAM of Xception model of cervical AC. a H&E; b Grad-CAM; c Guided Grad-CAM
Fig. 5.
H&E, Grad-CAM, and Guided Grad-CAM of benign images misclassified by the Xception model. a H&E; b Grad-CAM; c Guided Grad-CAM
Discussion
Main Findings
This study was aimed at identifying cervical malignancies and providing diagnostic interpretability. Our results demonstrated that pre-trained CNNs were able to distinguish cervical cancer images from nonmalignant images accurately and effectively. Xception achieved the best performance in distinguishing both cervical SCC and AC from benign tissues, with an AUC of 0.974 and 0.958, respectively. The performance of CNNs falls between experienced and inexperienced pathologists. Moreover, Xception was able to provide the probability heatmap highlighting the malignant signatures, such as network pattern infiltrating and hyperchromatic nuclei with coarse chromatin.
Interpretation
Designing better network architectures has become a trend to improve the performance of CNN. It is well known that the performance of CNN can be significantly improved by increasing the number of stacked layers (also known as depth) [12]. Compared with AlexNet, the remaining three CNNs consist of more layers with a growing number of hyper-parameters. The growth of depth may be the reason why AlexNet gets worse performance than other CNNs in all classification tasks. Theoretically, the performance of CNNs improves along with the increasing volume of labeled image data [17]. All CNNs yielded different declines in performance when they were tasked with distinguishing AC from benign tissues, as AC is a relatively rare type of cervical cancer, making up only a small number of labeled images. Xception yielded less decline in performance than other CNNs when it was applied in tasks with a small number of labeled images. In addition to increasing the depth of CNN architecture, Xception utilizes necessary block, separable convolutions, which greatly narrow down the complexity, lessen the burden of operations, and allow CNNs to learn fewer parameters more efficiently. These results indicated that neural networks with more sophisticated designs might be suitable for tasks with a small number of labeled images.
Currently, the procedure of cervical cancer screening is based on a typical workflow, including HPV testing, cytology or PAP smear testing, colposcopy, and biopsy. Aside from HPV testing, correct diagnosis of the remaining three techniques is burdensome and very demanding on pathologists’ experience. However, in developing countries like China, more than half of the pathologists only have primary technical qualifications when they perform cervical cancer screening [18]. Thanks to the investigation of the graphic processing units (GPU) and improvements in CNN architectures, CNNs have been applied to facilitate the screening of cervical cancer. Cao et al. constructed an attention-guided CNN to classify cervical cytology images into normal/abnormal classes and achieved 95.08% accuracy and 95.83% sensitivity, which is comparable to the results of an experienced pathologist with 10 years of experience [19]. They also concluded that the average diagnostic time of CNN is 0.04 s per image, much quicker than the average time of pathologists (14.83 s per image). Chandran et al. developed CNNs to detect cervical cancer using colposcopy images. Constructed CNN exhibited sensitivity and specificity of 92.4 and 96.2%, respectively [20]. However, the application of CNNs on cervical biopsy received much less attention than other screening techniques, such as cytology and colposcopy. Only one study constructed AlexNet, one of the simplest CNN architectures, to distinguish malignant from benign pathologic cervical images, achieving an accuracy of 93.33% [21]. In the present study, the constructed CNNs not only achieved a diagnostic performance slightly weaker than that of pathologists with more than 10 years of experience but were also significantly faster in diagnostic time than human experts (CNN: 0.03 s per image; human: 8.74 per image).
Strengths and Limitations
The strength of the study lies in that we could provide diagnostic interpretability by Grad-CAM and Guided Grad-CAM. The main drawback of CNNs’ application is that for many pathologists, the function of CNNs remains a black box, leading to less convincing evidence and more exaggerated expectations [22]. The ability to understand which parts of a given image led a convent to its final classification decision not only provide convicting evidence but also assist clinicians in finding high possible malignant area. Grad-CAM is highly class discriminative. Grad-CAM that aims to explain “malignant” could exclusively highlight the “malignant” regions. Guided Grad-CAM, which combined Grad-CAM and existing pixel-space gradient visualization (guided backpropagation) is not only class discriminative but also high resolution, which could highlight fine-grained details on the image [23]. Therefore, the major advantage of Grad-CAM is its ability to investigate and explain classification mistakes. As shown in Fig. 3, Grad-CAM and Guided Grad-CAM successfully highlighted the regions characterized by malignant changes, including large and hyperchromatic nuclei. Meanwhile, Grad-CAM can reflect why an image is misclassified by CNNs. However, Grad-CAM also has its limitations. The main drawback of Grad-CAM is the inability to localize multiple occurrences of the same class [24]. Even for single-object images, Grad-CAM failed to localize the entire region of the object. In this work, Grad-CAM could not highlight all regions with malignant changes in one image with multiple regions of malignant changes (see Figs. 3 and 4), which may limit its application in teaching inexperienced pathologists.
The shortage of labeled histological images is the first limitation of our study. Theoretically, the larger amount of labeled images, the better the performance of CNNs for a specific cancer classification task. Due to the laborious work both in image acquirement and annotation, we spent one year collecting and labeling 8496 H&E images from a total of 229 cervical specimens. Moreover, there is no public open-access database containing the histological image of cervical carcinoma. In recent years, whole slide image (WSI) has become a popular technique for pathologic digitalization [25]. The size of a WSI can reach 200 MB to 1 GB because it records the information of the entire tissue section, containing trillions of pixels [26]. The abundance of information per WSI provided sufficient data for establishing a diagnostic model for a cancer classification task. We plan to deploy the WSI technique in the future to obtain more labeled images to enrich the cervical cancer dataset.
The second limitation is that we only construct classification tasks for two histology subtypes of cervical cancer. Although these are the representative subtypes of cervical cancer, the rare types of cancer, such as neuroendocrine tumors or mesenchymal tumors, could not be distinguished accurately and efficiently by the present diagnostic tool. Future studies collecting images of rare types of cervical cancer are needed to make comprehensive cancer diagnostic systems possible.
Conclusion
In conclusion, our work demonstrates that a diagnostic tool with the excellent discriminative ability for binary classification tasks of histologic images could be achieved by using CNNs on a dataset composed of 8496 H&E images. Xception achieves 0.974 AUC for distinguishing cervical SCC and 0.958 AUC for distinguishing cervical AC from benign tissue. However, the performance of Xception is slightly weaker than experienced pathologists. By deploying Grad-CAM and Guided Grad-CAM, CNNs can provide diagnostic interpretability by highlighting the region of higher malignant probability. CNN could serve as a diagnostic tool to help pathologists to facilitate the diagnosis process of cervical cancer.
Supplementary Information
Below is the link to the electronic supplementary material.
Author Contribution
LYX and CF contributed equally to this research. LYX, CF, and WM devised the study plan. LYX, CF, SJJ, and HYL helped with data acquisition. LYX, CF, and SJJ wrote the draft manuscript. WM and CF supervised the research.
Funding
This research was financially supported by the Chongming district Innovation and Entrepreneurship Project (granted by Mei Wang).
Declarations
Ethics Approval and Consent to Participate
The study protocol was approved by the institutional review board of Xinhua hospital (CMEC-2022-KT-15). Written informed consent was waived by the institutional review boards owing to the retrospective study design. However, verbal informed consent was obtained from all patients.
Consent for Publication
The authors affirm that verbal informed consent for publication was obtained from all patients.
Conflict of Interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yi-xin Li and Feng Chen equally contributed to this study.
References
- 1.Arbyn M, et al. Estimates of incidence and mortality of cervical cancer in 2018: a worldwide analysis. Lancet Glob Health. 2020;8(2):e191–e203. doi: 10.1016/S2214-109X(19)30482-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gien LT, Beauchemin MC, Thomas G. Adenocarcinoma: a unique cervical cancer. Gynecol Oncol. 2010;116(1):140–146. doi: 10.1016/j.ygyno.2009.09.040. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, et al. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 4.Wu SY, Huang EY, Lin H. Optimal treatments for cervical adenocarcinoma. Am J Cancer Res. 2019;9(6):1224–1234. [PMC free article] [PubMed] [Google Scholar]
- 5.Sun H, et al. Computer-aided diagnosis in histopathological images of the endometrium using a convolutional neural network and attention mechanisms. IEEE J Biomed Health Inform. 2020;24(6):1664–1676. doi: 10.1109/JBHI.2019.2944977. [DOI] [PubMed] [Google Scholar]
- 6.Albayrak A, et al. A whole-slide image grading benchmark and tissue classification for cervical cancer precursor lesions with inter-observer variability. Med Biol Eng Comput. 2021;59(7–8):1545–1561. doi: 10.1007/s11517-021-02388-w. [DOI] [PubMed] [Google Scholar]
- 7.Pouliakis A, et al. Using classification and regression trees, liquid-based cytology and nuclear morphometry for the discrimination of endometrial lesions. Diagn Cytopathol. 2014;42(7):582–591. doi: 10.1002/dc.23077. [DOI] [PubMed] [Google Scholar]
- 8.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 9.Ehteshami Bejnordi B, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. Jama. 2017;318(22):2199–2210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huff, D.T., A.J. Weisman, and R. Jeraj, Interpretation and visualization techniques for deep learning models in medical imaging. Phys Med Biol, 2021. 66(4): p. 04tr01. [DOI] [PMC free article] [PubMed]
- 11.Krizhevsky, A., I. Sutskever, and G. Hinton, ImageNet classification with deep convolutional neural networks. Advances in neural information processing systems, 2012. 25(2).
- 12.Simonyan, K. and A. Zisserman, Very deep convolutional networks for large-scale image recognition. Computer Science, 2014.
- 13.He, K., et al., Deep residual learning for image recognition. IEEE, 2016.
- 14.Chollet, F., Xception: Deep learning with depthwise separable convolutions. IEEE, 2017.
- 15.Zhou, B., et al., Learning Deep features for discriminative localization. IEEE Computer Society, 2016.
- 16.Springenberg∗, J., et al., Striving for simplicity: the all convolutional net. eprint arxiv, 2014.
- 17.Litjens G, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 18.Di J, Rutherford S, Chu C. Review of the cervical cancer burden and population-based cervical cancer screening in China. Asian Pac J Cancer Prev. 2015;16(17):7401–7407. doi: 10.7314/APJCP.2015.16.17.7401. [DOI] [PubMed] [Google Scholar]
- 19.Cao L, et al. A novel attention-guided convolutional network for the detection of abnormal cervical cells in cervical cancer screening. Med Image Anal. 2021;73:102197. doi: 10.1016/j.media.2021.102197. [DOI] [PubMed] [Google Scholar]
- 20.Chandran V, et al. Diagnosis of cervical cancer based on ensemble deep learning network using colposcopy images. Biomed Res Int. 2021;2021:5584004. doi: 10.1155/2021/5584004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu, M., et al., Automatic classification of cervical cancer from cytological images by using convolutional neural network. Biosci Rep, 2018. 38(6). [DOI] [PMC free article] [PubMed]
- 22.Adadi A, Berrada M. Peeking inside the black-box: a survey on Explainable Artificial Intelligence (XAI) IEEE Access. 2018;6:52138–52160. doi: 10.1109/ACCESS.2018.2870052. [DOI] [Google Scholar]
- 23.Selvaraju, R.R., et al. Grad-cam: Visual explanations from deep networks via gradient-based localization. in Proceedings of the IEEE international conference on computer vision. 2017.
- 24.Aditya, C., et al., Grad-CAM++: improved visual explanations for deep convolutional networks. arXiv 2018. arXiv preprint arXiv:1710.11063.
- 25.Jahn, S.W., M. Plass, and F. Moinfar, Digital pathology: advantages, limitations and emerging perspectives. J Clin Med, 2020. 9(11). [DOI] [PMC free article] [PubMed]
- 26.Ying X, Monticello TM. Modern imaging technologies in toxicologic pathology: an overview. Toxicol Pathol. 2006;34(7):815–826. doi: 10.1080/01926230600918983. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.