Abstract
Skin cancer is one of the primary causes of death globally, and experts diagnose it by visual inspection, which can be inaccurate. The need for developing a computer-aided method to aid dermatologists in diagnosing skin cancer is highlighted by the fact that early identification can lower the number of deaths caused by skin malignancies. Among computer-aided techniques, deep learning is the most popular for identifying cancer from skin lesion images. Due to their power-efficient behavior, spiking neural networks are attractive deep neural networks for hardware implementation. We employed deep spiking neural networks using the surrogate gradient descent method to classify 3670 melanoma and 3323 non-melanoma images from the ISIC 2019 dataset. We achieved an accuracy of 89.57% and an F1 score of 90.07% using the proposed spiking VGG-13 model, which is higher than the VGG-13 and AlexNet using less trainable parameters.
Keywords: Deep learning, Image analysis, Spiking neural networks, Skin lesion classification
Introduction
Skin cancer is one of the most prevalent types of cancer in the USA [1, 2]. It is estimated that every one out of five Americans can develop skin cancers in their life [3] and around 95,000 people in the USA are diagnosed with skin cancer every year, according to the estimate provided in [4].
Melanocytes are the cells that give the tan or brown color to the skin [5]. Uncontrolled growth of melanocytes results in melanoma, which is less common but one of the most dangerous types of skin cancer in the world [5]. One million people in the USA carry melanoma [6]. Twenty people die in the USA daily due to melanoma [7]. Around 197,700 Americans will be diagnosed with melanoma in 2022, according to the study in [6]. Three million Americans are diagnosed with non-melanoma cancers like basal cell carcinoma (BCC) and squamous cell carcinoma (SCC) [4]. Ninety-nine percent of people survive if melanoma is detected early [6]. These facts emphasize the need for early detection of skin cancers.
Medical experts diagnose skin cancers by visual inspection, which can be time-consuming and inaccurate [8]. About 6% of skin cancers are correctly diagnosed as melanoma. The diagnostic accuracy of skin cancers can be improved using dermoscopy [8]. Dermoscopy assesses the colors and microstructures of the epidermis, the dermo-epidermal intersection, by illuminating the image, which the exposed eye cannot see [9]. Accurate diagnosis of skin cancers using dermoscopy is highly dependent on the skills and experience of the examiner due to similarities in color, shape, and size of skin lesions. These differences can result in different diagnoses by different examiners. These dermoscopy issues stress the importance of developing computer-aided methods for early diagnosing skin cancers.
Computer-aided method for diagnosing skin cancer involves image acquisition, pre-processing, segmentation, feature extraction, and classification [10, 11]. Image segmentation and classification are complex due to varying skin colors, size, the texture of lesions, and artifacts such as hairs, air bubbles, shadows, markers, and color calibration charts [12, 13].
This paper will focus on classifying skin lesion images as melanoma and non-melanoma. In recent years machine learning (ML) methods have been used for the automated detection and classification of skin cancer images improving the accuracy by 15–20% [14]. Deep learning, the most popular machine learning method for image recognition and classification [15], has also shown excellent performance in skin lesion detection and classification [16]. The following section will review literature that uses deep learning to classify skin cancers.
Related Work
Nasr-Esfahani et al. used a convolutional neural network (CNN) to classify melanoma. The convolutional neural network comprising two convolution layers trained on 170 skin lesions achieved an average accuracy of 81% [17]. Ali and Al-Marzouqi proposed CNN based melanoma classification network based on LightNet [18] in [19]. The proposed architecture, trained using fewer parameters, achieved an accuracy of 81.6% on the ISIC 2016 dataset. Esteva et al. proposed the Deep CNN pertained on ImageNet [20] in [21] for the classification of skin cancers. Esteva et al. compared the binary classification of the proposed Deep CNN model with 21 certified dermatologists. Esteva et al. considered two binary classification cases, keratinocyte carcinomas versus benign seborrheic keratoses; and malignant melanomas versus benign nevi. The proposed model trained on 129,450 clinical images achieved an accuracy of 72%. Dorj et al. combined support vector machines (SVM) and a deep neural network for the classification of skin cancers into four classes, squamous cell carcinoma (SCC), actinic keratosis (AK), basal cell carcinoma (BCC), and melanoma in [22]. Dorj et al. classified skin cancers into four classes using the features extracted by the pre-trained AlexNet. The proposed method trained on 3753 dermoscopic images achieved a melanoma classification accuracy of 94%. Harangi et al. proposed an ensemble method comprising of AlexNet, VGGNet [24], and GoogleNet [25] for the classification of skin cancers into three classes. The proposed ensemble method trained on 2000 images from ISIC 2017 [26] achieved an average AUC of 0.84 and an average accuracy of 84.8%. Kalouche et al. classified the skin lesion image as melanoma and non-melanoma using logistic regression, deep neural network, and pre-trained VGG-16 in [27]. Pre-trained VGG-16 model classified the skin lesion images as melanoma with a balance accuracy score of 78%. Rezvantalab et al. compared the classification performance of different deep learning algorithms with the dermatologists in [28]. Rezvantalab et al. classified the skin cancers into eight categories using Google’s Inception v3 [29], InceptionResNet v2 [30], ResNet 152 [31], DenseNet 201 [32]. All these networks were trained on 10,135 dermoscopy images. The classification accuracy was, on average, 11% higher than the dermatologists. Albahar proposed a two-layer CNN network based on a novel regularizer for classifying skin lesion images in [33]. The proposed model achieved an AUC of 0.77, 0.85, and 0.86 for nevus versus melanoma, SK versus melanoma, and solar lentigo (SL) cases. Mahbod et al. used three pre-trained deep learning models, AlexNet, VGG16, and ResNet-18, for extracting features. Three SVM classifiers were trained separately on the features extracted using pre-trained networks. Fi on classifying skin lesion images into three classes, malignant melanoma, SK, and benign nevus, using SVM in [34]. The final classification score was achieved by averaging the results of all three classifiers. The proposed method was tested by classifying 150 skin lesion images from ISIC into three classes, malignant melanoma, SK, and benign nevus. Skin lesions were accurately classified as melanoma with an average AUC of 0.833. The overall average AUC was 0.9069. Yu et al. presented an image recognition system for classifying melanoma images in [35]. Features were extracted using deep CNN. Then, the fisher vector encoding was used to generate representative features given to the SVM classifier using a chi-squared kernel to classify melanoma images. This method was intended to solve the problems caused by the limited training data. The proposed system trained on ISIC 2016 dataset achieved an accuracy of 86.81%. Majtner et al. proposed a method for increasing the accuracy of melanoma detection by extracting features using the combination of deep neural network and linear discriminant analysis in [36]. Extracted features were then used for training five different classifiers. The proposed model trained on 900 images from ISIC 2016 dataset achieved the highest accuracy of 86%. Jojoa Acosta et al. proposed a two-stage method for classifying melanoma images in [37]. Mask and region-based convolutional network was used for cropping the region of interest from skin images. ResNet152 was trained on these images for classification purposes in the next stage. The proposed model trained on 2742 images from ISIC 2016 achieved the maximum balance accuracy of 87.2%. Sagar and Jacob presented a method for classifying skin lesion images as melanoma and non-melanoma in [38]. ResNet 50 with deep transfer learning trained on 3600 lesion images for the ISIC dataset achieved an accuracy of 93.5% and an F1 score of 85%.
We can achieve good skin lesion accuracy using deep convolution neural networks or combining different deep models. However, they are best suited for deployment on portable devices due to resource consumption problems [39]. Neurons in ANN process and transmit analog information, different from the functioning of biological neurons in the brain [40]. Spiking neural networks work similarly to the brain’s biological neurons by processing and transmitting information using a sequence of spikes at different points in time [41]. SNNs are suited for event-driven hardware operations due to their biologically plausible nature [40]. Neurons in SNN process information only after receiving input spike trains. The event-driven hardware consumes less power as the part of the network not driven by input spikes can be power gated [42].
Spiking neural networks have shown good performance in pattern recognition applications. Kheradpisheh et al. used the shallow architecture of spike-timing-dependent plasticity (STDP) based SNN for object recognition in [43]. Escobar et al. proposed SNN-based framework for action recognition in [44]. Liu and Yue used STDP bases SNN to classify MNIST digits in [45]. SNN has been used for odor classification in [46]. SNNs have also been used for classification purposes in biomedical applications. Lobov et al. combined spiking neurons and artificial neurons for the feature extraction and classification of surface electromyography (sEMG) in [47]. Spiking neurons were used for feature extraction, while classification was done through artificial neurons. The proposed hybrid model using spiking neurons and artificial neurons achieved higher classification accuracy than other state-of-the-art ANN models. Tan et al. proposed an emotion classification system on a multimodal dataset using evolving SNN (eSNN) in [48]. Tan et al. also developed an encoding method for mapping facial features to spikes using population coding. Kasabov et al. used evolving SNN for the prediction of stroke in individuals and achieved an overall accuracy of 94% in [49]. Ghosh-Dastidar and Adeli [50] proposed SNN for epilepsy and seizure detection using electroencephalogram (EEG) signals achieved the highest accuracy of 92.5% in [49]. Zhou et al. classified skin lesion images using STDP-based SNN. The proposed method achieved an accuracy of 83.8% on the ISIC 2018 dataset in [39]. Zhou et al. also showed that the accuracy of the proposed model was improved to 87.8% using the feature selection method.
Training of SNNs is not straightforward. SNNs process information in the form of spikes which are discrete values. We cannot use standard backpropagation algorithms for training as the spikes generated by SNN are non-differentiable. SNNs can be trained using either supervised or unsupervised learning methods. STDP is the most popular unsupervised training method due to its energy-on-chip efficient learning. However, their performance is not comparable to supervised training methods and cannot be used with deep architectures. Syed et al. showed that the deep SNN trained using surrogated gradient descent, a supervised learning method, achieved better accuracy with few timesteps. We also propose deep SNN architecture trained using the surrogate gradient descent learning method for skin lesion classification.
The main contributions of our work are:
We proposed a deep spiking neural network (Spiking VGG-13) with an integrate and fire neuron (IF) model trained using surrogate gradient descent and fewer timesteps to classify skin lesions.
We compared the performance of our model with VGG-13, AlexNet.
We also compared the performance of our model with other spiking models, such as ResNet-18 and VGG-13, built using spikingjelly [52].
Overview of the Proposed Method
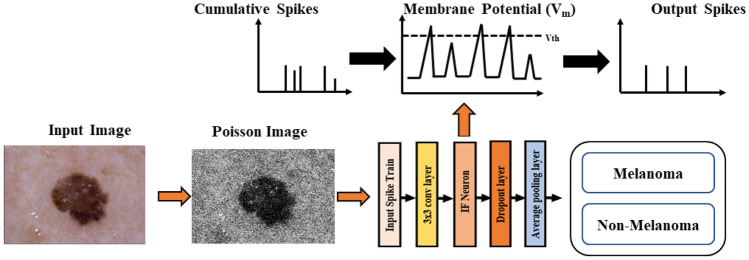
Figure 1 presents an overview of the proposed method. The input image is converted into spikes using Poisson distribution, and these input spikes trains are passed through convolution layers. When a integrate and fire neuron (IF) reaches a certain threshold, it spikes, and the potential of that IF neuron is reset.
Fig. 1.
Skin cancer classification using SNN
Integrate and Fire (IF) Neuron Model
This article employs a basic integrate and fire (IF) neuron model to reduce the number of possible hyperparameters. For time-independent classification problems, the IF neuron model is adequate. On the other hand, more complicated neuron models, such as least leaky integrate and fire (LIF), may be required for more complex and time-dependent tasks.
Equation (1) describes the internal mechanism of the membrane potential [53] of the IF neuron.
| 1 |
where C denotes the membrane capacitance and in the synaptic input current. The input is an integration of weighted pre-neuron spikes , with representing the connecting synaptic weights. When the membrane potential of the neuron reaches the specified threshold value , the output spikes are generated, and the is decreased to the resting potential .
Ledinauskas et al. [54] has changed Eq. (1) to incorporate discrete time increments by substituting the differential equation with the difference equation:
| 2 |
The output spikes are = 1 if > u and 0 otherwise. The neuron’s membrane potential resets once the output spikes are generated. Ledinauskas et al. [54] have considered two kinds of reset: reset to zero = which is known as hard reset [55] and on the other hand soft reset is represented by subtraction [56, 57]. It has been claimed that soft reset diminishes information loss and enhances ANN-SNN conversion [58, 59]. Ledinauskas et al. [54] have found that training a shallow model that contains several layers with a soft reset permits them to acquire more suitable classification performance than models with a hard reset. On the other hand, a hard reset shows better performance in deep models. Ledinauskas et al. [54] have stated that the advantage of a hard reset for deeper models may be connected to the accumulation of inaccuracies owing to surrogate gradient since the temporal record of the membrane potential is lost after a hard reset.
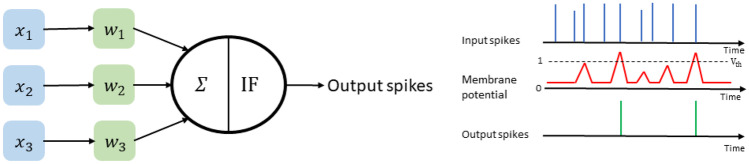
The generation of output spikes in the IF neuron model is summarized in Fig. 2. The input spikes (pre-synaptic neurons) are controlled by inter-connecting synaptic weights to generate output spikes (post-synaptic neurons). IF neurons integrate the inputs from all neurons to develop membrane potential ; if the membrane potential crosses the threshold value , IF neuron generates an output spike, then membrane potential resets to the starting value.
Fig. 2.
Output Spike generation mechanism of neurons in the IF neuron model
Proposed Deep Spiking Neural Network for Melanoma Classification
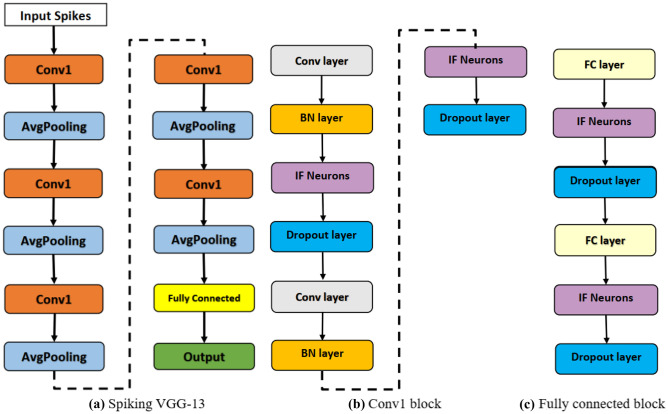
The proposed spiking VGG-13 model for the classification of skin cancer is presented in Fig. 3. Spiking VGG-13 consists of five conv1 blocks, one fully connected (FC) block, and average pooling layers followed by conv1 blocks. Each conv1 block consists of two convolution layers followed by a batch normalization (BN) layer and an IF neurons layer. FC block consists of two fully connected layers followed by IF neurons and a dropout layer. 3 3 convolution layers were used for learning features from skin lesion images. The dropout layer was used to tackle the overfitting problem as discussed in [51], and average pooling was used to decrease the input dimension size. BN layers were used to perform regularization that helps the network converge faster. The final classification is performed using fully connected layers.
Fig. 3.
Proposed architecture for skin cancer classification
Training Deep Spiking Neural Networks Using Surrogate Gradient Decent
Initially, assuming a single neuron and exploring how to train the neuron to emit a particular spike train for a specific stimulus [54]. This study has stated an error function in this fashion: the energy is a time integral of power which is equivalent to the error current increased by the membrane voltage of the neuron . Thus, the loss function is determined as follows:
| 3 |
where T is the entire spike train time, such a loss function has no significant problems with the spiking nonlinearity’s non-differentiability because the gradient of the output O(t) is invalid and ignored by [60], using the function . When the output spike pulse is missing , the loss function specified in (3) has specific necessary properties: the loss value decreases with increasing membrane voltage, allowing the output spike train to occur. However, when the output spike is absent, the loss increases with the decrease in membrane voltage . The drawback of this loss function is that it may produce both positive and negative values, and values close to zero can be produced when the membrane voltage is close to zero, even if the output spikes vary from the expected ones. On the other hand, the desired values for the weights equal to the gradient loss are zero.
Likewise, [54] indicated a broader loss employing the van Rossum distance.
| 4 |
| 5 |
where (5) depicts a convolution operation. For instance, If the number of spike trains exceeds a particular threshold value, only one neuron may be employed for classification purposes. Additionally, Zenke and Ganguli [60] have used the convolutional kernel for the whole period T in (4) and provided the loss function below for classification purposes:
| 6 |
where denotes the ground-truth labels and denotes a Heaviside step function. In (4), the gradient of the loss function may be determined as follows:
| 7 |
Equation (3) may be employed as a substitute and
| 8 |
instead of (6) combined with a customized rule
| 9 |
Backpropagation has been employed to train SNNs by replacing the gradient of a non-differentiable spike with an alternate surrogate gradient [61]. The simple conversion , however, this kind of gradient causes issues when used to deeper layers of SNN. When a neuron’s membrane voltage in a hidden layer is close to zero, gradient descent with the surrogate gradient allows the potential to rise or drop with almost equal probability. The membrane voltage remains close to zero for a longer duration. To avoid this problem, Ledinauskas et al. [54] have adapted the conversion rule as:
| 10 |
when the membrane voltage reaches nearly zero, is reduced: The value of f(0) is smaller than . According to mathematical findings, the actual method of the function is not important. Ledinauskas et al. [54] have changed the conversion rule as:
| 11 |
The surrogate gradient size is characterized by , and the gradient thickness is represented by a. The previous section shows that the term is inadequate for deeper SNNs. Apart from severe instances when the membrane voltage approaches the threshold value, large values of a make the surrogate gradient frequently relatively small. As a result, it is plausible that an ideal intermediate value of a exists. Note that we assume the gradients of spikes and hard reset SNN neurons to be zero. After resetting, the membrane voltage gradient is unaffected by previous membrane voltage values ; the history of the neuron’s membrane potential is forgotten.
Experiments and Results
In this section, we evaluated the performance of the proposed spiking VGG-13 model for melanoma versus non-melanoma classification. We also compared the performance of proposed spiking VGG-13 models with state-of-the-art deep neural networks and the spiking models from spikingjelly [52].
Dataset
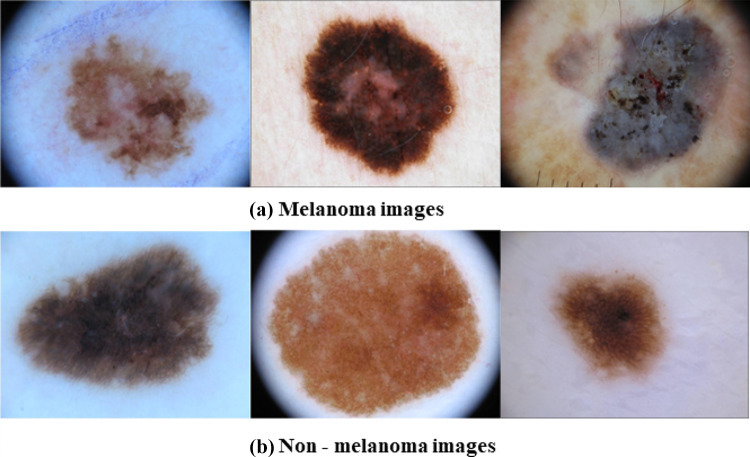
This work focuses on distinguishing melanoma from non-melanoma skin lesion images and uses images from the ISIC 2019 dataset, which is highly imbalanced. We only used 3670 melanoma and 3323 non-melanoma images from the ISIC dataset to address the class imbalance. A few cases of melanoma and non-melanoma from ISIC 2019 used for training spiking VGG-13 are shown in Fig. 4.
Fig. 4.
Skin lesion images from ISIC 2019
Experimental Settings
The proposed model used IF neuron models with the threshold value of 1. We used the random split method to divide the ISIC dataset into training, validation, and test set; 70% of the dataset was used as a training set, 15% of the dataset was used as a validation set, and 15% of the dataset is used as a test set. The model was trained for 100 epochs with a learning rate of 0.003 using an SGD optimizer and a batch size of 8. The number of time steps used was 30. Fully connected layers had 1024 neurons, while output layers had just two neurons, as this is a binary classification problem. 2 2 filter size was used in the average pooling layer. Cross entropy was used as a loss function. Analysis was performed in PyTorch.
Evaluation Metrics
Developing an automated skin lesion detection system aims to increase the diagnostic accuracy of skin cancers. In this work, we are classifying melanoma, the most dangerous type of skin cancer, using SNN. The metrics used for evaluating this work are Accuracy, F1 score, sensitivity, specificity, and precision. We can calculate these metrics using the following equations:
| 12 |
| 13 |
| 14 |
| 15 |
| 16 |
where TP is the number of true positives, TN is the number of true negatives, FP is the number of false positives, and FN is the number of false negatives.
Results
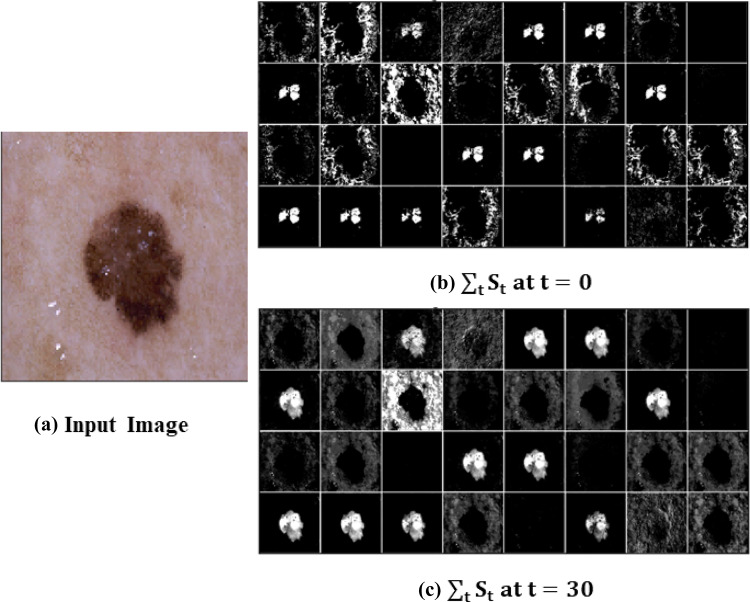
Figure 5 presents the melanoma features extracted using spiking VGG-13 at different time steps. It is challenging to visualize deep spiking neural networks; we are just showing the features extracted after the first layer. Spiking VGG-13 is learning the complex features over time for accurate melanoma classification. Features are more prominent at the time, t =30, compared to the features extracted at t = 0.
Fig. 5.
Melanoma features visualization using spiking VGG-13
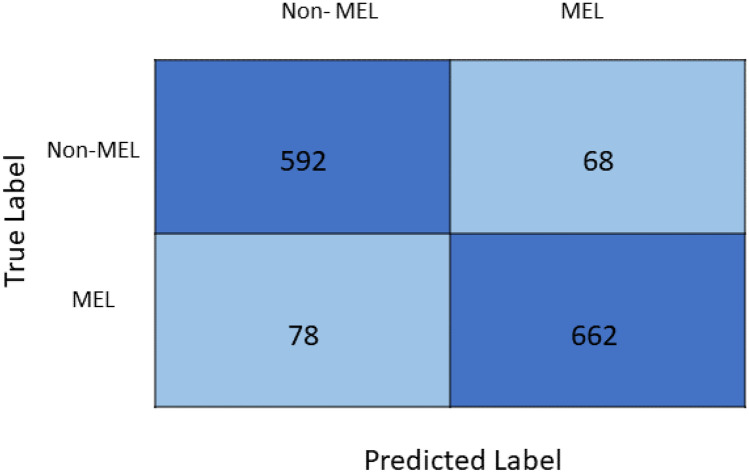
Figure 6 presents the confusion matrix of the proposed spiking VGG-13 model. Five hundred ninety-two out of 660 cases were correctly classified as non-melanoma cases, while 68 non-melanoma cases were misclassified as melanoma. Six hundred sixty-two out of 740 cases were correctly classified as melanoma, while 78 melanoma cases were misclassified as non-melanoma.
Fig. 6.
Confusion matrix of the proposed spiking VGG-13 model
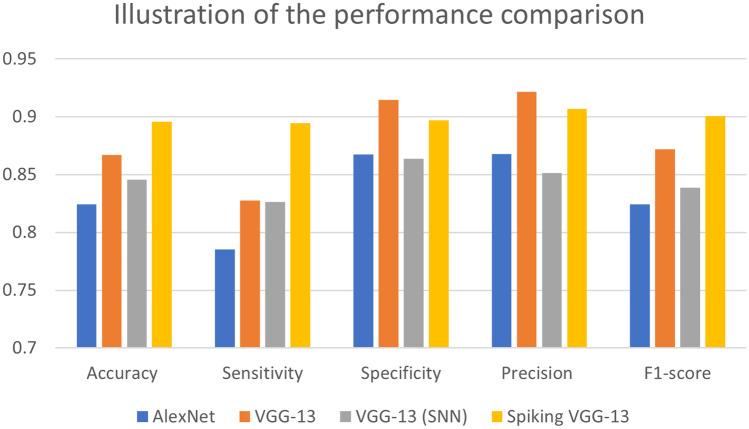
We compared the performance of the proposed spiking VGG-13 model with AlexNet, VGG-13, and VGG-13 (SNN) in Table 1. Spiking VGG-13 achieved the best accuracy of 0.8957 compared to other models. Spiking VGG-13 also had the highest F1-score of 0.9007 and sensitivity value of 0.8946. The specificity and precision values of the VGG-13 model were slightly higher than our proposed model. VGG-13 achieved a specificity of 0.9148 and a precision of 0.9215, while our proposed model spiking VGG-13 achieved a specificity score of 0.8970 and a sensitivity score of 0.9068. Spiking VGG-13 gave the best performance overall in comparison with the other models. Figure 7 illustrates the performance comparison of spiking VGG-13 with other models.
Table 1.
Performance comparison of the proposed model with the state-of-the-art
| Model | Accuracy | Sensitivity | Specificity | Precision | F1 score |
|---|---|---|---|---|---|
| AlexNet | 0.8243 | 0.7853 | 0.8675 | 0.8679 | 0.8245 |
| VGG-13 | 0.8671 | 0.8277 | 0.9148 | 0.9215 | 0.8721 |
| VGG-13(SNN) [52] | 0.8457 | 0.8265 | 0.8639 | 0.8515 | 0.8388 |
| Spiking VGG-13 (ours) | 0.8957 | 0.8946 | 0.8970 | 0.9068 | 0.9007 |
Bold values show the best results for accuracy, sensitivity, specificity, precision, and F1 score
Fig. 7.
Graphical illustration of the performance comparison
In the end, we compared the number of parameters of spiking VGG-13 with deep neural network counterparts. Spiking VGG-13 has the minimum number of trainable parameters compared to AlexNet and VGG-13, as shown in Table 2. We need to train only 14,879,040 parameters of spiking VGG-13 which were relatively less than the trainable parameters of AlexNet and VGG-13.
Table 2.
Performance comparison of the proposed model with the state-of-the-art
| Model | No. of parameters |
|---|---|
| AlexNet | 57,012,034 |
| VGG-13 | 128,959,042 |
| Spiking VGG-13 (ours) | 14,879,040 |
Discussion
Spiking neural networks are known to outperform conventional neural network architecture in computational complexity, time, and energy resources, which makes them ideal for hardware deployments such as neuromorphic kits by intel and other manufacturers. The advantage hails from the fact that dermatologists spend a lot of time diagnosing the correct type of cancer cell; added on top is the fact that the diagnosis rate is low. SNN helps dermatologists leverage the efficient neural network for timely and highly accurate diagnosis. Based on the aforementioned advantages, SNN for cancer diagnosis is suitable for clinical settings.
Conclusion
An early skin cancer diagnosis can significantly increase the survival rate of skin cancer patients worldwide. There is a strong need to develop automated skin cancer detection systems that can assist dermatologists in diagnosing skin cancers. We presented an SNN-based cancer detection system famous for its hardware implementation due to its power-efficient nature. The proposed Spiking VGG-13 gave the best performance compared to the other state-of-the-art deep neural network used for skin cancer detection.
Author Contribution
Project conception: Syed Qasim Gilani, Tehreem Syed, Oge Marques. Drafting of the manuscript: Syed Qasim Gilani, Tehreem Syed. Design and modeling: Syed Qasim Gilani, Tehreem Syed, Muhammad Umair. Experiments: Syed Qasim Gilani. Supervision: Oge Marques.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Declarations
Conflict of Interest
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Syed Qasim Gilani, Email: [email protected].
Tehreem Syed, Email: [email protected].
Muhammad Umair, Email: [email protected].
Oge Marques, Email: [email protected].
References
- 1.Gery P Guy Jr, Cheryll C Thomas, Trevor Thompson, Meg Watson, Greta M Massetti, and Lisa C Richardson. Vital signs: melanoma incidence and mortality trends and projections–united states, 1982–2030. MMWR. Morbidity and mortality weekly report, 64 (21):591, 2015. [PMC free article] [PubMed]
- 2.Gery P Guy Jr, Steven R Machlin, Donatus U Ekwueme, and K Robin Yabroff. Prevalence and costs of skin cancer treatment in the us, 2002- 2006 and 2007- 2011. American journal of preventive medicine, 48 (2):183–187, 2015. [DOI] [PMC free article] [PubMed]
- 3.Robert S Stern. Prevalence of a history of skin cancer in 2007: results of an incidence-based model. Archives of dermatology, 146 (3):279–282, 2010. [DOI] [PubMed]
- 4.Howard W Rogers, Martin A Weinstock, Steven R Feldman, and Brett M Coldiron. Incidence estimate of nonmelanoma skin cancer (keratinocyte carcinomas) in the us population, 2012. JAMA dermatology, 151 (10): 1081–1086, 2015. [DOI] [PubMed]
- 5.https://tinyurl.com/39sj38eb. Accessed Apr 2022.
- 6.https://tinyurl.com/yfb73knk. Accessed Apr 2022.
- 7.Rebecca L Siegel, Kimberly D Miller, Hannah E Fuchs, and Ahmedin Jemal. Cancer statistics, 2022. CA: a cancer journal for clinicians, 2022. [DOI] [PubMed]
- 8.Harold Kittler, H Pehamberger, K Wolff, and MJTIO Binder. Diagnostic accuracy of dermoscopy. The lancet oncology, 3 (3): 159–165, 2002. [DOI] [PubMed]
- 9.Ashfaq A Marghoob, Lucinda D Swindle, Claudia ZM Moricz, Fitzgeraldo A Sanchez Negron, Bill Slue, Allan C Halpern, and Alfred W Kopf. Instruments and new technologies for the in vivo diagnosis of melanoma. Journal of the American Academy of Dermatology, 49 (5): 777–797, 2003. [DOI] [PubMed]
- 10.Afsaneh Jalalian, Syamsiah Mashohor, Rozi Mahmud, Babak Karasfi, M Iqbal B Saripan, and Abdul Rahman B Ramli. Foundation and methodologies in computer-aided diagnosis systems for breast cancer detection. EXCLI journal, 16: 113, 2017. [DOI] [PMC free article] [PubMed]
- 11.Fan Haidi, Xie Fengying, Li Yang, Jiang Zhiguo, Liu Jie. Automatic segmentation of dermoscopy images using saliency combined with otsu threshold. Computers in biology and medicine. 2017;85:75–85. doi: 10.1016/j.compbiomed.2017.03.025. [DOI] [PubMed] [Google Scholar]
- 12.Md Kamrul Hasan, Lavsen Dahal, Prasad N Samarakoon, Fakrul Islam Tushar, and Robert Martí. Dsnet: Automatic dermoscopic skin lesion segmentation. Computers in Biology and Medicine, 120: 103738, 2020. [DOI] [PubMed]
- 13.Korotkov Konstantin, Garcia Rafael. Computerized analysis of pigmented skin lesions: a review. Artificial intelligence in medicine. 2012;56(2):69–90. doi: 10.1016/j.artmed.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Konstantina Kourou, Themis P Exarchos, Konstantinos P Exarchos, Michalis V Karamouzis, and Dimitrios I Fotiadis. Machine learning applications in cancer prognosis and prediction. Computational and structural biotechnology journal, 13: 8–17, 2015. [DOI] [PMC free article] [PubMed]
- 15.LeCun Yann, Bengio Yoshua, Hinton Geoffrey. Deep learning. nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 16.Michel Fornaciali, Micael Carvalho, Flávia Vasques Bittencourt, Sandra Avila, and Eduardo Valle. Towards automated melanoma screening: Proper computer vision & reliable results. arXiv preprint arXiv:1604.04024, 2016.
- 17.Ebrahim Nasr-Esfahani, Shadrokh Samavi, Nader Karimi, S Mohamad R Soroushmehr, Mohammad H Jafari, Kevin Ward, and Kayvan Najarian. Melanoma detection by analysis of clinical images using convolutional neural network. In 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pages 1373–1376. IEEE, 2016. [DOI] [PubMed]
- 18.Chengxi Ye, Chen Zhao, Yezhou Yang, Cornelia Fermüller, and Yiannis Aloimonos. Lightnet: A versatile, standalone matlab-based environment for deep learning. In Proceedings of the 24th ACM international conference on Multimedia, pages 1156–1159, 2016.
- 19.Aya Abu Ali and Hasan Al-Marzouqi. Melanoma detection using regular convolutional neural networks. In 2017 International Conference on Electrical and Computing Technologies and Applications (ICECTA), pages 1–5. IEEE, 2017.
- 20.Alex Krizhevsky, Ilya Sutskever, and Geoffrey E Hinton. Imagenet classification with deep convolutional neural networks. Advances in neural information processing systems, 25, 2012.
- 21.Andre Esteva, Brett Kuprel, Roberto A Novoa, Justin Ko, Susan M Swetter, Helen M Blau, and Sebastian Thrun. Dermatologist-level classification of skin cancer with deep neural networks. nature, 542 (7639): 115–118, 2017. [DOI] [PMC free article] [PubMed]
- 22.Dorj Ulzii-Orshikh, Lee Keun-Kwang, Choi Jae-Young, Lee Malrey. The skin cancer classification using deep convolutional neural network. Multimedia Tools and Applications. 2018;77(8):9909–9924. doi: 10.1007/s11042-018-5714-1. [DOI] [Google Scholar]
- 23.Balazs Harangi, Agnes Baran, and Andras Hajdu. Classification of skin lesions using an ensemble of deep neural networks. In 2018 40th annual international conference of the IEEE engineering in medicine and biology society (EMBC), pages 2575–2578. IEEE, 2018. [DOI] [PubMed]
- 24.Karen Simonyan and Andrew Zisserman. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556, 2014.
- 25.Christian Szegedy, Wei Liu, Yangqing Jia, Pierre Sermanet, Scott Reed, Dragomir Anguelov, Dumitru Erhan, Vincent Vanhoucke, and Andrew Rabinovich. Going deeper with convolutions. In Proceedings of the IEEE conference on computer vision and pattern recognition, pages 1–9, 2015.
- 26.Noel CF Codella, David Gutman, M Emre Celebi, Brian Helba, Michael A Marchetti, Stephen W Dusza, Aadi Kalloo, Konstantinos Liopyris, Nabin Mishra, Harald Kittler, et al. Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (isbi), hosted by the international skin imaging collaboration (isic). In 2018 IEEE 15th international symposium on biomedical imaging (ISBI 2018), pages 168–172. IEEE, 2018.
- 27.Simon Kalouche, Andrew Ng, and John Duchi. Vision-based classification of skin cancer using deep learning. 2015, conducted on Stanfords Machine Learning course (CS 229) taught, 2016.
- 28.Amirreza Rezvantalab, Habib Safigholi, and Somayeh Karimijeshni. Dermatologist level dermoscopy skin cancer classification using different deep learning convolutional neural networks algorithms. arXiv preprint arXiv:1810.10348, 2018.
- 29.Christian Szegedy, Vincent Vanhoucke, Sergey Ioffe, Jon Shlens, and Zbigniew Wojna. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE conference on computer vision and pattern recognition, pages 2818–2826, 2016.
- 30.Christian Szegedy, Sergey Ioffe, Vincent Vanhoucke, and Alexander A Alemi. Inception-v4, inception-resnet and the impact of residual connections on learning. In Thirty-first AAAI conference on artificial intelligence, 2017.
- 31.Kaiming He, Xiangyu Zhang, Shaoqing Ren, and Jian Sun. Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition, pages 770–778, 2016.
- 32.Gao Huang, Zhuang Liu, Laurens Van Der Maaten, and Kilian Q Weinberger. Densely connected convolutional networks. In Proceedings of the IEEE conference on computer vision and pattern recognition, pages 4700–4708, 2017.
- 33.Marwan Ali Albahar Skin lesion classification using convolutional neural network with novel regularizer. IEEE Access. 2019;7:38306–38313. doi: 10.1109/ACCESS.2019.2906241. [DOI] [Google Scholar]
- 34.Amirreza Mahbod, Gerald Schaefer, Chunliang Wang, Rupert Ecker, and Isabella Ellinge. Skin lesion classification using hybrid deep neural networks. In ICASSP 2019-2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), pages 1229–1233. IEEE, 2019.
- 35.Zhen Yu, Jiang Xudong, Zhou Feng, Qin Jing, Ni Dong, Chen Siping, Lei Baiying, Wang Tianfu. Melanoma recognition in dermoscopy images via aggregated deep convolutional features. IEEE Transactions on Biomedical Engineering. 2018;66(4):1006–1016. doi: 10.1109/TBME.2018.2866166. [DOI] [PubMed] [Google Scholar]
- 36.Tomáš Majtner, Sule Yildirim-Yayilgan, and Jon Yngve Hardeberg. Optimised deep learning features for improved melanoma detection. Multimedia Tools and Applications, 78 (9): 11883–11903, 2019.
- 37.Mario Fernando Jojoa Acosta, Liesle Yail Caballero Tovar, Maria Begonya Garcia-Zapirain, and Winston Spencer Percybrooks. Melanoma diagnosis using deep learning techniques on dermatoscopic images. BMC Medical Imaging, 21 (1): 1–11, 2021. [DOI] [PMC free article] [PubMed]
- 38.Abhinav Sagar and Dheeba Jacob. Convolutional neural networks for classifying melanoma images. bioRxiv, pages 2020–05, 2021.
- 39.Zhou Qian, Shi Yan, Zhenghua Xu, Ruowei Qu, Guizhi Xu. Classifying melanoma skin lesions using convolutional spiking neural networks with unsupervised stdp learning rule. IEEE Access. 2020;8:101309–101319. doi: 10.1109/ACCESS.2020.2998098. [DOI] [Google Scholar]
- 40.Sengupta Abhronil, Ye Yuting, Wang Robert, Liu Chiao, Roy Kaushik. Going deeper in spiking neural networks: Vgg and residual architectures. Frontiers in neuroscience. 2019;13:95. doi: 10.3389/fnins.2019.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ponulak Filip, Kasinski Andrzej. Introduction to spiking neural networks: Information processing, learning and applications. Acta neurobiologiae experimentalis. 2011;71(4):409–433. doi: 10.55782/ane-2011-1862. [DOI] [PubMed] [Google Scholar]
- 42.Zhanping Chen, Mark Johnson, Liqiong Wei, and W Roy. Estimation of standby leakage power in cmos circuit considering accurate modeling of transistor stacks. In Proceedings. 1998 International Symposium on Low Power Electronics and Design (IEEE Cat. No. 98TH8379), pages 239–244. IEEE, 1998.
- 43.Saeed Reza Kheradpisheh, Mohammad Ganjtabesh, Simon J Thorpe, and Timothée Masquelier. Stdp-based spiking deep convolutional neural networks for object recognition. Neural Networks, 99: 56–67, 2018. [DOI] [PubMed]
- 44.Maria-Jose Escobar, Guillaume S Masson, Thierry Vieville, and Pierre Kornprobst. Action recognition using a bio-inspired feedforward spiking network. International journal of computer vision, 82 (3): 284–301, 2009.
- 45.Liu Daqi, Yue Shigang. Fast unsupervised learning for visual pattern recognition using spike timing dependent plasticity. Neurocomputing. 2017;249:212–224. doi: 10.1016/j.neucom.2017.04.003. [DOI] [Google Scholar]
- 46.Anup Vanarse, Josafath Israel Espinosa-Ramos, Adam Osseiran, Alexander Rassau, and Nikola Kasabov. Application of a brain-inspired spiking neural network architecture to odor data classification. Sensors, 20 (10): 2756, 2020. [DOI] [PMC free article] [PubMed]
- 47.Lobov Sergey, Mironov Vasiliy, Kastalskiy Innokentiy, Kazantsev Victor. A spiking neural network in semg feature extraction. Sensors. 2015;15(11):27894–27904. doi: 10.3390/s151127894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clarence Tan, Gerardo Ceballos, Nikola Kasabov, and Narayan Puthanmadam Subramaniyam. Fusionsense: Emotion classification using feature fusion of multimodal data and deep learning in a brain-inspired spiking neural network. Sensors, 20 (18): 5328, 2020. [DOI] [PMC free article] [PubMed]
- 49.Kasabov Nikola, Feigin Valery, Hou Zeng-Guang, Chen Yixiong, Liang Linda, Krishnamurthi Rita, Othman Muhaini, Parmar Priya. Evolving spiking neural networks for personalised modelling, classification and prediction of spatio-temporal patterns with a case study on stroke. Neurocomputing. 2014;134:269–279. doi: 10.1016/j.neucom.2013.09.049. [DOI] [Google Scholar]
- 50.Ghosh-Dastidar Samanwoy, Adeli Hojjat. Improved spiking neural networks for eeg classification and epilepsy and seizure detection. Integrated Computer-Aided Engineering. 2007;14(3):187–212. doi: 10.3233/ICA-2007-14301. [DOI] [Google Scholar]
- 51.Syed Tehreem, Kakani Vijay, Cui Xuenan, Kim Hakil. Exploring optimized spiking neural network architectures for classification tasks on embedded platforms. Sensors. 2021;21(9):3240. doi: 10.3390/s21093240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei Fang, Yanqi Chen, Jianhao Ding, Ding Chen, Zhaofei Yu, Huihui Zhou, Yonghong Tian, and other contributors. Spikingjelly. https://github.com/fangwei123456/spikingjelly, 2020. Accessed: YYYY-MM-DD.
- 53.Wulfram Gerstner, Werner M Kistler, Richard Naud, and Liam Paninski. Neuronal dynamics: From single neurons to networks and models of cognition. Cambridge University Press, 2014.
- 54.Eimantas Ledinauskas, Julius Ruseckas, Alfonsas Juršėnas, and Giedrius Buračas. Training deep spiking neural networks. arXiv preprint arXiv:2006.04436, 2020.
- 55.Peter U Diehl, Daniel Neil, Jonathan Binas, Matthew Cook, Shih-Chii Liu, and Michael Pfeiffer. Fast-classifying, high-accuracy spiking deep networks through weight and threshold balancing. In 2015 International joint conference on neural networks (IJCNN), pages 1–8. ieee, 2015.
- 56.Andrew S Cassidy, Paul Merolla, John V Arthur, Steve K Esser, Bryan Jackson, Rodrigo Alvarez-Icaza, Pallab Datta, Jun Sawada, Theodore M Wong, Vitaly Feldman, et al. Cognitive computing building block: A versatile and efficient digital neuron model for neurosynaptic cores. In The 2013 International Joint Conference on Neural Networks (IJCNN), pages 1–10. IEEE, 2013.
- 57.Peter U Diehl, Bruno U Pedroni, Andrew Cassidy, Paul Merolla, Emre Neftci, and Guido Zarrella. Truehappiness: Neuromorphic emotion recognition on truenorth. In 2016 international joint conference on neural networks (ijcnn), pages 4278–4285. IEEE, 2016.
- 58.Bing Han, Gopalakrishnan Srinivasan, and Kaushik Roy. Rmp-snn: Residual membrane potential neuron for enabling deeper high-accuracy and low-latency spiking neural network. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pages 13558–13567, 2020.
- 59.Rueckauer Bodo, Lungu Iulia-Alexandra, Yuhuang Hu, Pfeiffer Michael, Liu Shih-Chii. Conversion of continuous-valued deep networks to efficient event-driven networks for image classification. Frontiers in neuroscience. 2017;11:682. doi: 10.3389/fnins.2017.00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zenke Friedemann, Ganguli Surya. Superspike: Supervised learning in multilayer spiking neural networks. Neural computation. 2018;30(6):1514–1541. doi: 10.1162/neco_a_01086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Emre O Neftci, Hesham Mostafa, and Friedemann Zenke. Surrogate gradient learning in spiking neural networks. IEEE Signal Processing Magazine, 36: 61–63, 2019.