Abstract
The rapid spread of coronavirus disease (COVID-19) has become a worldwide pandemic and affected more than 15 million patients reported in 27 countries. Therefore, the computational biology carrying this virus that correlates with the human population urgently needs to be understood. In this paper, the classification of the human protein sequences of COVID-19, according to the country, is presented based on machine learning algorithms. The proposed model is based on distinguishing 9238 sequences using three stages, including data preprocessing, data labeling, and classification. In the first stage, data preprocessing’s function converts the amino acids of COVID-19 protein sequences into eight groups of numbers based on the amino acids’ volume and dipole. It is based on the conjoint triad (CT) method. In the second stage, there are two methods for labeling data from 27 countries from 0 to 26. The first method is based on selecting one number for each country according to the code numbers of countries, while the second method is based on binary elements for each country. According to their countries, machine learning algorithms are used to discover different COVID-19 protein sequences in the last stage. The obtained results demonstrate 100% accuracy, 100% sensitivity, and 90% specificity via the country-based binary labeling method with a linear support vector machine (SVM) classifier. Furthermore, with significant infection data, the USA is more prone to correct classification compared to other countries with fewer data. The unbalanced data for COVID-19 protein sequences is considered a major issue, especially as the US’s available data represents 76% of a total of 9238 sequences. The proposed model will act as a prediction tool for the COVID-19 protein sequences in different countries.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11517-021-02412-z.
Keywords: COVID-19 protein sequences, Conjoint triad (CT), Machine learning algorithms, Support vector machine (SVM)
Introduction
Resistance to the coronavirus is still a challenge due to the limited information available about this virus. The available cases of coronavirus protein sequences encouraged research on the taxonomic classification of the COVID-19 virus [1]. The early differentiation of this virus resolved the massive spread, especially in dynamic population growth. Many factors focused on the probabilities of developing the virus in patients, such as age, hygienic behaviors, location, environment, and health status [2]. The World Health Organization (WHO) reported that the COVID-19 virus, which originated in China, can spread in many countries [3]. The WHO also confirmed that this virus, referred to as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), emerged in bats and was transmitted to humans [4]. This means that exposure to the specific wild environment is providing more rapid infections. The infection risk of COVID-19 is controlled by accurate quarantine schedules because there are no specific drugs or vaccines for this virus [5]. COVID-19 has had an important influence on society’s financial and social aspects, causing a critical need to reduce the spread of this virus. Generally, the viral analysis of protein sequence has a good indication of its responsibility for pharmacological interactions.
Moreover, the classification task of the virus protein sequences by machine learning algorithms supports treatment plans for COVID-19 [6]. Additionally, the National Center for Biotechnology Information (NCBI) [7] provided a list of genomic datasets targeting COVID-19 to aid in coronavirus variant analysis as a public resource. To combat this virus, the genetic features of COVID-19 should be well known. It is a single-stranded RNA virus with a diameter ranging from 65 to 125 nm, consisting of around 27–32 kb [8].
Generally, protein–protein interactions (PPI) are employed for studying virus proteins regarding their replications and interactions [9]. The traditional methods for PPI are related to laboratory tests such as tandem affinity purification [10], yeast two-hybrid system [11], and mass spectrometry [12]. However, these tests are suffered from high costs, time consumption, human errors, and restricted coverage. The computational algorithms have struggled to predict protein sequences. Therefore, PPI’s computational prediction [13] is a more effective solution than traditional methods. The computational prediction models [14–17] are developed to extract the features from amino acids with diverse accuracy levels. Recent studies proved that the CT method [18] had undergone a high prediction accuracy for PPI. The successful representation of the CT method is based on the adjoining influence of the relative three kinds of amino acids [19].
Zhang et al. [20] presented the protein sequence analysis for host identification of COVID-19 and studied the similarity between COVID-19 and HIV (human immunodeficiency virus). Randhawa et al. [6] presented machine learning algorithms of DNA sequences to classify COVID-19 as a beta coronavirus. Qiang et al. [21] proposed the observation model of protein sequences of COVID-19 to study its genomic evolution via machine learning algorithms. Zhou et al. [22] suggested the PPI network for recognizing the candidate drugs for COVID-19. The prediction of PPI for HIV, SARS, and pandemic influenza A (H1N1) virus [23] identified by amino acid composition is shown in previous research [24]. Host-virus PPI proposed by the co-immunoprecipitation method has also been shown in [25].
On the other hand, some researchers presented an effective model for determining mutation locations in the DNA sequences of this coronavirus [26]. Desautels et al. [27] suggested a machine learning–based prediction model of antibody structures qualified for attacking the COVID-19 receptor-binding domain. Recently, Dey et al. [28] focused on the train of interactions between COVID-19 and human proteins using machine learning approaches to encourage COVID-19 drug discovery.
The numbers of infected cases and fatality rates associated with COVID-19 change from country to country. Toyoshima et al. [29] confirmed that there had been no explanation of the reasons for these genomic variations based on infected countries. Also, the temperature divergences in different countries [30] created mutations of COVID-19 protein sequences and have prompted researchers to study geographical variations from a bioinformatics perspective. Therefore, the COVID-19 protein sequence classification based on the country is a significant way to track the virus. Transmission of this virus through different countries might affect the potential for mutations and severity of COVID-19. Each country has created a database for COVID-19 protein sequences, which is an important research point for virus recognition through classification algorithms. This means that the COVID-19 protein sequences have categories equal to the number of endemic countries. Hence, the COVID-19 treatment based on antivirals and vaccines will adjust according to virus recognition based on the affected country.
In this study, the proposed model investigated the CT method for the classification of COVID-19 human protein sequences considering their countries through machine learning techniques. This is the initial processing related to infection areas of COVID-19, which has been recorded recently. Also, the proposed model explored the different forms of COVID-19 protein sequences and their relationships to mortality rates in 27 countries. This study aims to promote the growth control of COVID-19 based on protein sequences and discover other mutation types of this pandemic that will support as a tool for the drug production of this virus in the health sector.
Materials and methods
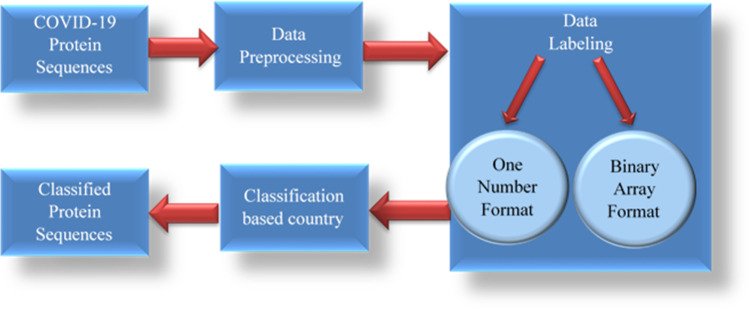
The proposed block diagram designed procedure series including data preprocessing, data labeling, and classification algorithms to categorize the COVID-19 countries, as shown in Fig. 1.
Fig. 1.
Block diagram of the classification of COVID-19 protein sequences based on the country
Dataset description
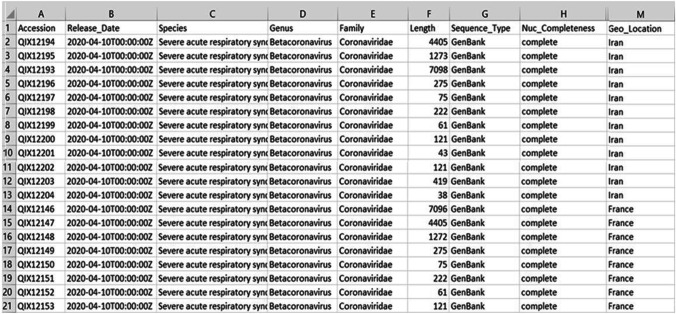
The used dataset for SARS-Cov-2 (COVID-19) was constructed by the NCBI virus [7]. This dataset contains 9238 sequences, each sequence was identified using the accession number and containing information such as protein sequence as a FASTA file, geographical location, and protein sequence length. There are two forms in the dataset. In the first form, comma-separated values (CSV) files include the accession numbers of protein sequences and other information, such as species, genus, family, length, sequence type, and geographical location (Geo_location), as shown in Fig. 2. The maximum sequence length found in this dataset is QIX12193, which has 7098 amino acids. The minimum sequence length found in this dataset is YP_009725312, which has 13 amino acids. In the second form, FASTA files contain the accessions and the protein sequences, as shown in Fig. 3. In the bioinformatics domain, the FASTA format is an arrangement of protein sequences using the alignment software package based on single-letter codes described by the European Bioinformatics Institute (EBI). The FASTA format is used to find the similarities between protein sequences.
Fig. 2.
Data samples of COVID-19 protein sequences in CSV format that include some information such as accession number, release date, species, genus, family, length, sequence type, and geographical location for each protein sequence
Fig. 3.
Data samples of COVID-19 protein sequences in FASTA format
Data preprocessing stage by conjoint triad (CT) method
Generally, the protein sequences consist of twenty amino acids. The frequency of every three adjacent amino acids is recognized as a feature (a triad). Thus, the extracted features in each protein sequence are 203 = 8000. It is more difficult to analyze protein sequences as raw data by machine learning approaches [31]. In the data preprocessing phase, classification of the amino acid side chains into seven classes ({AGV}, {ILFP}, {YMTS}, {HNQW}, {RK}, {DE}, {C}) is applied by the conjoint triad (CT) method [18]. The CT method is used to transform the amino acids in each sequence into numbers according to their volumes and dipoles. Most approaches of PPI [32–34] are implemented by the CT method to reduce the extracted features to 73 = 343 according to seven classes.
Mathematically, we consider a protein sequence P with length L is P = P1P2P3 ……. PL and all the adjacent three amino acids in P are P1P2P3, P2P3P4, ….. P3P4P5,..., PL-2PL-1PL. Then, the CT feature for protein sequence represents normalized frequency where CT is defined as follows.
| 1 |
where is the frequency of each triad.
Molecular modeling and density-functional theory approaches used Gaussian03 system [35] for calculating volumes and dipoles of the side chains of the amino acids. The Gaussian03 system [35] is used as a computational chemistry software to avoid traditional computations’ cost. The functions of the Gaussian03 system are listed in the following:
(1) Calculation of one- and two-electron integrals over any general contracted Gaussian functions. The basic functions can either be Cartesian Gaussians or pure angular momentum functions, and a variety of basis sets are stored in the program and can be requested by name.
(2) Transformation of the atomic orbital (AO) integrals to the molecular orbital basis.
(3) Molecular mechanics calculations.
(4) Correlation energy calculations using Møller-Plesset perturbation theory with high accuracy.
(5) Speed up the density functional theory (DFT) calculations.
This CT method converts the protein sequences to numbers from zero to seven. The classification of the amino acids based on their side-chain dipoles and volumes is shown in Table 1.
Table 1.
Amino acid classification according to dipoles and volumes using the CT method
| Class number | Dipole scale | Volume scale | Amino acids |
|---|---|---|---|
| 1 | - | - | A, G, V |
| 2 | - | + | I, L, F, P |
| 3 | + | + | Y, M, T, S |
| 4 | + + | + | H, N, Q, W |
| 5 | + + + | + | R, K |
| 6 | + ' + ' + ' | + | D, E |
| 7 | + | + | C |
Some explanations of the symbols are found in Table 1.
• Dipole scale: (-), dipole < 1.0/( +), 1.0 < dipole < 2.0/(+ +), 2.0 < dipole < 3.0 /(+ + +), dipole > 3.0 / (+ ' + ' + '), dipole > 3.0 with opposite orientation
• Volume scale: (-), volume < 50;/( +), volume > 50
• Cysteine (C) amino acids are separated from class 3 because of their ability to form disulfide bonds
As shown in Table 2, the new amino acid class was added to the coding with class number zero and amino acid labeled with (X). The protein sequence “ALGCERQSKXTP” would be represented as “121,765,435,032” according to eight amino acid classes. The sample of COVID-19 protein sequences after conversion is displayed in Fig. 4. The CT method is employed as encoding each protein sequence using the continuous three amino acid frequency distribution extracted from 7 letters to reduce the characters of amino acids.
Table 2.
Amino acid coding according to eight classes
| Class number | Amino acids |
|---|---|
| 0 | X (unknown amino acid) |
| 1 | A, G, V |
| 2 | I, L, F, P |
| 3 | Y, M, T, S |
| 4 | H, N, Q, W |
| 5 | R, K |
| 6 | D, E |
| 7 | C |
Fig. 4.
Protein sequences after conversion using eight amino acid classes
After converting the protein sequences to numbers, an array of eight elements is filled with each amino acid class’s number for each sequence in the dataset. Finally, the amino acids’ frequencies are normalized by dividing each frequency by the length of the sequence to avoid bias to the taller sequences.
Data labeling stage
The classification of protein sequences based on the country is implemented on the COVID-19 dataset. The countries’ names are labeled in Table 3. There are 27 unique countries found in this dataset after removing the cities.
Table 3.
Code numbers of countries
| Country number | Country name |
|---|---|
| 0 | Australia |
| 1 | Belgium |
| 2 | Brazil |
| 3 | Spain |
| 4 | Colombia |
| 5 | Finland |
| 6 | France |
| 7 | India |
| 8 | Iran |
| 9 | Iraq |
| 10 | Israel |
| 11 | Italy |
| 12 | Japan |
| 13 | Malaysia |
| 14 | Nepal |
| 15 | Nigeria |
| 16 | Pakistan |
| 17 | Peru |
| 18 | Philippines |
| 19 | South Korea |
| 20 | China |
| 21 | Sweden |
| 22 | Taiwan |
| 23 | Thailand |
| 24 | Tunis |
| 25 | USA |
| 26 | Vietnam |
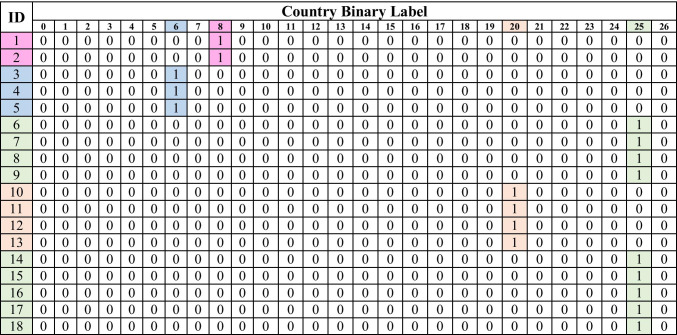
To prepare these countries as labels for the classifier. There are two methods as follows:
Each country has a number from 0 to 26, and each record has only one number as a label according to its country. A sample of the labeled sequences is displayed in Table 4.
Each record has an array of 27 binary elements in which 1 represents its country; otherwise, it is zero, as shown in Table 5.
Table 4.
One number of format samples

Table 5.
Binary array format samples
Data division
After the data labeling stage, all labeled data is randomly divided with a proportion of 8:2 into a training set and a testing set. In machine learning algorithms, the value of training and testing is a significant factor in deciding the performance level [36]. If the features and the label have a high correlation, the training–testing ratio is 50%–50%. This means that half of the data is used for training and half is used for testing. If there is a fear of success slipping away, the training rate may be increased. In previous researches, the training–testing ratio varies according to the data structure. It is based on the trials and errors by changing the training and testing ratio to achieve a high accuracy rate [37]. By many experiments, it was found that 80% of training and 20% of testing achieved a high accuracy rate in this proposed model. Less or more than the selected ratio in the training and testing process is not preferred because the test results will be adversely affected. Subsequently, the classification stage was taught using the training data and the final results were calculated using the testing data.
Classification stage
After converting the protein sequences of COVID-19 to numbers that belong to eight amino acid classes, six different classifiers were sequentially used for predicting the protein sequence country. Each amino acid class was considered a feature in this classification model. One number format and binary array format are the two methods of labeling the predicted class (country). The six classifiers are carried out by linear regression (LR) [38], K-nearest neighbor (KNN) [39] using various numbers of neighbors, support vector machine (SVM) [40] using different kernel functions, naive Bayesian (NB) [41], decision tree (DT) [42], and random forest (RF) [43] using different numbers of estimators. The classifier structure’s main objective is to reduce the learning complexity for the accurate examination of unknown samples.
Machine learning algorithms
These six machine learning algorithms can be applied to the classification problem of COVID-19 protein sequences according to the country as the following:
The LR classifier [38] is supported by the relationship between the dependent and independent variables using a linear separating line. The best separating line is called the regression line and represented by a linear equation Y = a *X + b, where Y is a dependent variable, a is a slope, X is an independent variable, and b is an intercept. The values a and b are derived based on reducing the sum of squared difference of distance between data points and regression line.
The KNN classifier [39] focused on distance measurement by the density distribution that was not related to decision boundary calculations. A class of their closest neighbors grades the KNN classifier based on K values. In this paper, the selected K values were 20, 50, 150, and 201 to attain the best K for results accuracy.
The SVM classifier [40] is based on plotting each data item as a point in n-dimensional space (where n is the number of features) with the value of each feature being the value of a particular coordinate (where coordinates are support vectors). In this paper, the SVM classifier investigated decision boundary calculations by four types of kernel functions, such as linear, sigmoid, polynomial, and radial basis function (RBF), to choose the effective function for discrimination.
The NB classifier [41] is based on the probability theorem and maximum likelihood calculation. This classifier assumes that the presence of a particular feature in a class is unrelated to the presence of any other feature.
The DT classifier [42] is based on data separation into groups according to low entropy measures. It splits the data into two or more homogeneous sets. This is done based on the most significant attributes/independent variables to make as distinct groups as possible.
The RF classifier [43] is based on large numbers of DT methods and a high correlation between two trees. The forest chooses the classification having the most votes (over all the trees in the forest). If the number of cases in the training set is N, then a sample of N cases is taken at random but with replacement. This sample will be the training set for growing the tree. If there are M input variables, a number m < < M is specified such that at each node, m variables are selected at random out of the M and the best split on these m is used to split the node. The value of m is held constant during forest growth. Each tree is grown to the largest extent possible. There is no pruning.
Performance metrics
The performance metrics for the proposed model are based on a multi-label confusion matrix and receiver operating characteristic (ROC) curve to evaluate COVID-19 classification algorithms according to each country’s protein sequence.
The multi-label confusion matrix is a common method for recording results in multi-class classification problems because it is possible to identify the relations between the classifier outputs and the true ones. The multi-label confusion matrix of all classification algorithms is based on the percentage of accuracy, sensitivity, and specificity that refer to the performance of each classifier. Accuracy is calculated as the rate of correctly classified sequences to the whole number of sequences in the protein data. Sensitivity is calculated as the rate of positives that are correctly identified sequences. Specificity is calculated as the rate of negatives that are correctly rejected. Three parameters are shown as the following equations (Eqs. 2–4).
| 2 |
| 3 |
| 4 |
where TP is true positive (correctly classified), TN is true negative (correctly rejected), FP is false positive (incorrectly classified), and FN is false negative (incorrectly rejected).
In this proposed model, the results covered the accuracy, sensitivity, and specificity rates for six algorithms based on two different labeling methods to assess the usefulness of the prediction model for COVID-19 according to country. It is a preliminary study to verify its accuracy in predicting COVID-19 disease according to protein sequences. The performance is calculated by taking the average of the accuracy rates obtained from each classification process. Therefore, the accuracy rate is a critical indicator in the training and testing process and classification success. In machine learning algorithms, if the testing data increases, the accuracy is expected to decrease [36].
The ROC curve [37] is another method to estimate the classification performance of machine learning algorithms. It represented the true positive rate (sensitivity) in the function of the false positive rate (specificity) for different cut-off points. Each point on the ROC curve showed a sensitivity/specificity pair corresponding to a particular decision threshold.
Experimental results
The dataset is separated into 80% for the training database and 20% for the testing database to perform the COVID-19 classification according to the country under the Python computational environment. The training–testing ratio is selected according to data correlation and performance criteria to achieve high algorithm accuracy [36]. It was found that the performance criteria can be maximized when the training data is greater than the testing data [37].
The CT method [18] is used to encode protein sequences to numbers according to seven classes, which have acted as feature extraction. Each feature group is estimated by the tenfold cross-validation test. There are two methods of class labeling used for the prediction of the COVID-19 country. The inclusive comparison between the six machine learning approaches is investigated on COVID-19 protein sequences. For the KNN classifier, the adjustable factor is the K value while for the SVM classifier, it is the kernel function, and it is an estimator number for the RF classifier. Based on twelve forms from six classifiers, the classification results for the one-number labeling method are illustrated in Table 6.
Table 6.
Performance comparison of different classifiers for the one number labeling method
| Classifiers | Accuracy% | Specificity% | Sensitivity% |
|---|---|---|---|
| LR | 77.7 | 75 | 80 |
| KNN, k = 20 | 77.2 | 72 | 88 |
| KNN, k = 50 | 77.7 | 74 | 88 |
| KNN, k = 150 | 77.7 | 75 | 88 |
| KNN, k > = 201 | 77.7 | 74 | 88 |
| SVM, Kernel: RBF | 78.0 | 76 | 88 |
| SVM, Kernel: linear | 77.2 | 71 | 88 |
| SVM, Kernel: sigmoid | 74.4 | 70 | 80 |
| SVM, Kernel: polynomial | 78.2 | 79 | 88 |
| NB | 13.2 | 45 | 30 |
| DT | 79.5 | 82 | 89 |
| RF, estimators = 10 | 79.0 | 81 | 89 |
Entries in bold indicate the best results
The preferable results confirmed that the DT classifier achieved an accuracy of 79.5%, a specificity of 82%, and a sensitivity of 89% for the one number labeling method. For the one-number labeling method, the worst classifier provided by NB achieved an accuracy of 13.2%, a specificity of 45%, and a sensitivity of 30%.
Based on twelve forms from six classifiers, the results of COVID-19 protein sequence–assisted detection for the binary labeling method are illustrated in Table 7. For the binary array method, the best classifier provided by linear SVM achieved an accuracy of 100%, sensitivity of 100%, and specificity of 90%.
Table 7.
Performance comparison of different classifiers for the binary array labeling method
| Classifiers | Accuracy% | Specificity% | Sensitivity% |
|---|---|---|---|
| LR | 100 | 86 | 100 |
| KNN, k = 20 | 93.4 | 81 | 98 |
| KNN, k = 50 | 95.7 | 86 | 98 |
| KNN, k = 150 | 99.8 | 88 | 100 |
| KNN, k > = 201 | 100 | 89 | 100 |
| SVM, Kernel: RBF | 99.6 | 87 | 98 |
| SVM, Kernel: linear | 100 | 90 | 100 |
| SVM, Kernel: sigmoid | 95.2 | 85 | 95 |
| SVM, Kernel: polynomial | 99.2 | 87 | 100 |
| NB | 99.8 | 86 | 99 |
| DT | 97.0 | 82 | 97 |
| RF, estimators = 10 | 96.5 | 85 | 98 |
Entries in bold indicate the best results
In contrast, linear SVM, KNN, and LR classifiers achieved an accuracy of 100%, as well as the linear SVM classifier, achieved a specificity of 100% for the binary array labeling method. The best sensitivity is 100% for LR, KNN (K > = 201), and SVM (linear, polynomial) classifiers. For the binary array labeling method, the worst classifier provided by KNN with K = 20 achieved an accuracy of 93.4%, specificity of 81%, and sensitivity of 89%.
Additionally, the best K value of ≥ 50 is obtained with 77.7% accuracy for the one number labeling method. The best K value of ≥ 201 is obtained with 100% accuracy for the binary array labeling method. The choice of K value has a slight impact on the classification accuracy for the one number labeling method. Still, it has a powerful impact on the classification accuracy for the binary labeling method.
In the SVM classifier, the best kernel function was polynomial for the one number labeling method that obtained 78.2% accuracy. The best kernel function was linear for the binary array labeling method that obtained 100% accuracy. The SVM classifier’s accuracy is altered only by changing the kernel functions for the binary labeling method.
Figure 5 displays the ROC curve for the best classifier for two labeling methods. The area under the ROC curve (AUC) is a measure of how well a parameter can distinguish between classes. It can be noted that the binary array labeling method is an effective method for the classification of COVID-19 protein sequences according to the country.
Fig. 5.
Average ROC curve for the best classifier. a DT classifier based on the one number labeling method. b Linear SVM classifier based on the binary array labeling method
The relationship between the eight amino acid classes and the frequency of amino acid classes in each COVID-19 protein sequence by using all samples found in the dataset (9238 sequences) concerning the eight classes of amino acids is displayed in Fig. 6. This figure confirmed that the COVID-19 virus has a high record of amino acids provided in the second class that consists of isoleucine (I), leucine (L), phenylalanine (F), and proline (P) amino acids.
Fig. 6.
COVID-19 samples with respect to amino acids classes
Discussion
The analysis of virus genome sequence is a serious tool for early diagnosis and identifies the infected geographical areas. Coronavirus [44] is the longest RNA virus which is categorized into four genera including alpha, beta, gamma, and delta. The alpha and beta viruses are known to infect humans. Bats are a normal host of the corona but not the only animal allowed to transfer the infection to humans. The WHO proved that SARS-CoV-2 is a bat coronavirus that attacks human cells. Recently, COVID-19 is a novel virus related to SARS-CoV-2, which is leading to increased morbidity and mortality rates in several counties. The explosive growth of data and its availability indicates the emergence of new COVID-19 analysis by machine learning algorithms.
This paper investigated the existing state-of-the-art solutions proposed for machine learning algorithms to address pandemic challenges related to COVID-19 protein sequences. The main objective of this paper is to classify the COVID-19 protein sequences according to infected countries. Also, it can be used for forecasting the COVID-19 sequences in different countries. Altogether, the results revealed that the binary array labeling method with a linear SVM classifier had the highest performance for the classification of COVID-19 protein sequences from the 27 countries.
The limitations of the current work are related to the unbalanced protein sequences of COVID-19 among different countries. The recorded data for COVID-19 protein sequences is not enough for the classification process or early prediction of COVID-19 patients according to the infection area. Therefore, data of COVID-19 protein sequences are required for all countries. The countries that are reported the COVID-19 cases should provide a database of virus genome sequences in public sources to encourage primary researches.
Also, the number of features needs to be carefully chosen if applied to an extremely large number of countries and cities. Especially, it found that the fatality rates of COVID-19 differ between regions or cities in the same country [45]. In the future, this point will encourage the researchers to find the classification among COVID-19 protein sequences in regions or cities in the same country.
So far, the mortality rates related to COVID-19 change among countries according to many factors such as age distribution, virus genomic types, and genetic backgrounds. It is not clear about the reasons for the high mortality rates of COVID-19 in the USA. It was noted that each country has a unique structure of COVID-19 protein sequence. Thus, it can create the genomic COVID-19 database for every country that is used for a COVID-19 diagnosis.
On the other hand, the limitation of the current work is based on using accuracy, sensitivity, and specificity as evaluation metrics for COVID-19 classification. Therefore, the evaluation metrics should be expanded to classify the COVID-19 protein sequence in the future.
In the future, analysis of COVID-19 genomic sequence should extend to the classification between COVID and other infectious sequences as well as a study of genomic mutations in a 3D protein structure to support the clinical trials to find drugs. Finally, the fight against the COVID-19 pandemic with scientific weapons will continue by gathering a huge database for this virus, developing the contributions of machine learning algorithms to the COVID-19 sequences. To explore this COVID-19 virus, there are main targets including therapeutics, diagnosis, and prevention that need more attention from researchers around the world.
Conclusion
In this paper, a dataset of 9238 COVID-19 protein sequences is used to evaluate the proposed model’s capability to predict the country of the protein sequences. The proposed model extracted suitable features from the protein sequences by replacing the amino acid characters in each sequence with the eight amino acid class normalized frequencies. After that, the model performed six different classifiers to predict the country of the virus protein sequence.
This model indicated that the classification using the binary array labeling method has more promising results than the one number labeling method. Hence, the preliminary proposed model elucidated the utility of the country binary array labeling method for discriminating COVID-19 protein sequences within an appropriate time using a linear SVM classifier.
The better classification accuracy was observed in US protein sequences compared with different countries because of their high data records, approximately 7020 records from 9238 records (76% from all occurrences). It is interesting to note that unbalanced data have a low impact on the COVID-19 classification results. This leads to increased misclassifications on the testing set because all countries’ COVID-19 sequences except the USA occupied only 24% of all sequences. Furthermore, the classifiers biased to US records compared with the countries with a small number of occurrences.
Finally, this proposed model could be expanded to large stored data of COVID-19 protein sequences and could then develop prediction algorithms for unbalanced sequences within several countries. Also, the proposed algorithm will expand to calculate further evaluation criteria such as precision to ensure the protein classification of COVID-19 according to country.
Supplementary Information
Below is the link to the electronic supplementary material.
Biographies
Dr. Heba M. Afify
had completed her B.Sc, M.Sc, and Ph.D. degrees from the Biomedical Engineering department at Cairo University, Egypt. Her research areas are Biomedical Image Processing and currently working on Bioinformatics and Biomechanics. She is working as an associate professor in Egypt. She had authored many research papers in reputed international and national journals. She was a technical reviewer and editorial board member for several international journals in the biomedical engineering field.
Engr. Muhammad S. Zanaty
had completed his B.Sc degree from Ain Shams University, Program of Bioinformatics, Department of Information System, Faculty of Computer and Information Sciences, Egypt.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Heba M. Afify, Email: [email protected]
Muhammad S. Zanaty, Email: [email protected]
References
- 1.Kannan S, Shaik Syed Ali P, Sheeza A, Hemalatha K. COVID-19 (novel coronavirus 2019) – recent trends. SARS Eur Rev Med Pharmacol Sci. 2020;24(4):2006–2011. doi: 10.26355/eurrev_202002_20378. [DOI] [PubMed] [Google Scholar]
- 2.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munster VJ, Koopmans M, van Doremalen N, van Riel D, de Wit E. A novel coronavirus emerging in china - key questions for impact assessment. N Engl J Med. 2020;382(8):692–694. doi: 10.1056/NEJMp2000929. [DOI] [PubMed] [Google Scholar]
- 4.Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention (2019) Novel coronavirus (2019-nCoV), Wuhan, China (2019). https://www.cdc.gov/coronavirus/2019-nCoV/summary.html
- 6.Randhawa GS, Soltysiak MPM, El Roz H, de Souza CPE, Hill KA, Kari L. Machine learning using intrinsic genomic signatures for rapid classification of novel pathogens: COVID-19 case study. PLoS ONE. 2020;15(4):e0232391. doi: 10.1371/journal.pone.0232391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.NCBI virus: https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/virus?SeqType_s=Nucleotide&VirusLineage_ss=Severe%20acute%20respiratory%20syndrome%20coronavirus%202%20(SARS-CoV2),%20taxid:2697049. [dataset]
- 8.Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sen R, Nayak L, De RK. A review on host-pathogen interactions: classification and prediction. Eur J Clin Microbiol Infect Dis. 2016;35:1581–1599. doi: 10.1007/s10096-016-2716-7. [DOI] [PubMed] [Google Scholar]
- 10.Huang H, Alvarez S, Nusinow DA. Data on the identification of protein interactors with the Evening Complex and PCH1 in Arabidopsis using tandem affinity purification and mass spectrometry (TAP–MS) Data Brief. 2016;8:56–60. doi: 10.1016/j.dib.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mehla J, Caufield JH, Uetz P. Mapping protein-protein interactions using yeast two-hybrid assays. Cold Spring Harb Protoc. 2015;5:442–452. doi: 10.1101/pdb.prot086157. [DOI] [PubMed] [Google Scholar]
- 12.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415(6868):180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 13.Skrabanek L, Saini HK, Bader GD, Enright AJ. Computational prediction of protein-protein interactions. Mol Biotechnol. 2008;38(1):1–17. doi: 10.1007/s12033-007-0069-2. [DOI] [PubMed] [Google Scholar]
- 14.You ZH, Zhu L, Zheng CH, Yu HJ, Deng SP, Ji Z. Prediction of protein-protein interactions from amino acid sequences using a novel multi-scale continuous and discontinuous feature set. BMC Bioinform. 2014;15(Suppl 15):S9. doi: 10.1186/1471-2105-15-S15-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng J, Li D, Wu Y, Zou Q, Liu X. An empirical study of features fusion techniques for protein-protein interaction prediction. Curr Bioinform. 2016;11(1):899–901. doi: 10.2174/1574893611666151119221435. [DOI] [Google Scholar]
- 16.Wang Y, You Z, Li X, Chen X, Jiang T, Zhang J. PCVMZM: using the probabilistic classification vector machines model combined with a zernike moments descriptor to predict protein-protein interactions from protein sequences. Int J Mol Sci. 2017;18(5):1029. doi: 10.3390/ijms18051029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He H, Zhao J, Sun G (2019) Computational prediction of MoRFs based on protein sequences and minimax probability machine. BMC Bioinformatics 20(529). 10.1186/s12859-019-3111-z [DOI] [PMC free article] [PubMed]
- 18.Shen J, Zhang J, Luo X, Zhu W, Yu K, Chen K, Li Y, Jiang H. Predicting protein-protein interactions based only on sequences information. Proc Natl Acad Sci U S A. 2007;104(11):4337–4341. doi: 10.1073/pnas.0607879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H, Hu X. Accurate prediction of nuclear receptors with conjoint triad feature. BMC Bioinf. 2015;16(1):402. doi: 10.1186/s12859-015-0828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang C, Zheng W, Huang X, Bell EW, Zhou X, Zhang Y. Protein structure and sequence reanalysis of 2019-nCoV genome refutes snakes as its intermediate host and the unique similarity between its spike protein insertions and HIV-1. J Proteome Res. 2020;19(4):1351–1360. doi: 10.1021/acs.jproteome.0c00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Qiang X, Xu V, Fang G, Liu W-B, Kou Z. Using the spike protein feature to predict infection risk and monitor the evolutionary dynamic of coronavirus. Infect Dis Poverty. 2020;9(1):33. doi: 10.1186/s40249-020-00649-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F (2020) Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov 6(14). 10.1038/s41421-020-0153-3 [DOI] [PMC free article] [PubMed]
- 23.Girard MP, Tam JS, Assossou OM, Kieny MP. The 2009 A (H1N1) influenza virus pandemic: A review. Vaccine. 2010;28(31):4895–4902. doi: 10.1016/j.vaccine.2010.05.031. [DOI] [PubMed] [Google Scholar]
- 24.Alguwaizani S, Park B, Zhou X, Huang DS, Han K. Predicting interactions between virus and host proteins using repeat patterns and composition of amino acids. J Healthc Eng. 2018;2018:1391265. doi: 10.1155/2018/1391265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Golemis E, Adams PD (2005) Protein-protein interactions: a molecular cloning manual, 2nd edn. CSHL Press, New York
- 26.Isa Irawan M, Mukhlash I, Rizky A, RirisatiDewi A. Application of Needleman-Wunch Algorithm to identify mutation in DNA sequences of corona virus. J Phys Conf Ser. 2019;1218:012031. doi: 10.1088/1742-6596/1218/1/012031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Desautels T, Zemla A, Lau E, Franco M, Faissol D (2020) Rapid in silico design of antibodies targeting SARS-CoV-2 using machine learning and supercomputing. bioRxiv. 10.1101/2020.04.03.024885
- 28.Dey L, Chakraborty S, Mukhopadhyay A (2020) Machine learning techniques for sequence-based prediction of viral–host interactions between SARS-CoV-2 and human proteins. Biomed J. 10.1016/j.bj.2020.08.003 [DOI] [PMC free article] [PubMed]
- 29.Toyoshima Y, Nemoto K, Matsumoto S, Nakamura Y, Kiyotani K. SARS-CoV-2 genomic variations associated with mortality rate of COVID-19. J Hum Genet. 2020;65:1075–1082. doi: 10.1038/s10038-020-0808-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiebe A, Longbottom J, Gleave K, Shearer FM, Sinka ME, Massey NC, et al. Geographical distributions of African malaria vector sibling species and evidence for insecticide resistance. Malar J. 2017;16(58):1–10. doi: 10.1186/s12936-017-1734-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aghajanbaglo S, Moosavi S, Rahgozar M, Rahimi A (2014) Predicting protein-protein interactions based on rotation of proteins in 3D-space, The Second International Workshop on Parallelism in Bioinformatics (PBio 2014), as part of IEEE Cluster
- 32.Wang H, Wu P. Prediction of RNA-protein interactions using conjoint triad feature and chaos game representation. Bioengineered. 2018;9(1):242–251. doi: 10.1080/21655979.2018.1470721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Zhang L, Jia L, Ren Y, Yu G. Protein-protein interactions prediction using a novel local conjoint triad descriptor of amino acid sequences. Int J Mol Sci. 2017;18(11):2373. doi: 10.3390/ijms18112373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, Wang R, Wei Y, Gui Y. A novel conjoint triad auto covariance (CTAC) coding method for predicting protein-protein interaction based on amino acid sequence. Math Biosci. 2019;313:41–47. doi: 10.1016/j.mbs.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 35.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Zakrzewski VG, Montgomery JA, Stratmann RE, Burant JC, et al (2003) GAUSSIAN 03 (Gaussian, Pittsburgh, PA), Revision C.02
- 36.Kürsad Uçar M, Nour M, Sindi H, Polat K. The effect of training and testing process on machine learning in biomedical datasets. Math Probl Eng. 2020;2020(2836236):1–17. doi: 10.1155/2020/2836236. [DOI] [Google Scholar]
- 37.Witten IH, Frank E, and Hall MA (2011) Credibility: evaluating what’s been learned, in data mining: practical machine learning tools and techniques. Morgan Kaufmann, Burlington, pp 147–187
- 38.Dreiseitla S, Ohno-Machado L. Logistic regression and artificial neural network classification models: a methodology review. J Biomed Inform. 2002;35:352–359. doi: 10.1016/S1532-0464(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 39.Cunningham P, Delany SJ (2007) k-Nearest neighbour classifiers, Technical Report UCD-CSI-2007–4, 1–17
- 40.Evgeniou T, Pontil M (2001) Support vector machines: theory and applications, ACAI 1999: Machine Learning and Its Applications 249–257
- 41.Rish I (2001) An empirical study of the naive bayes classifier. In: IJCAI 2001 workshop on empirical methods in artificial intelligence, vol 3. IBM, New York, pp 41–46
- 42.Gutierrez-Rodríguez A, Martínez-Trinidad JF, García-Borroto M, Carrasco- Ochoa J. Mining patterns for clustering on numerical datasets using unsupervised decision trees. Knowl. Based Syst. 2015;82:70–79. doi: 10.1016/j.knosys.2015.02.019. [DOI] [Google Scholar]
- 43.Breiman L. Random Forests. Mach Learn. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 44.Bruns D, Kraguljac N, Bruns T. COVID- 19: facts, cultural considerations, and risk of stigmatization. J Transcult Nurs. 2020;31(4):1–7. doi: 10.1177/1043659620917724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Becerra-Flores M, Cardozo T. SARS-CoV-2 viral spike G614 mutation exhibits higher case fatality rate. Int J Clin Pract. 2020;00:e13525. doi: 10.1111/ijcp.13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.