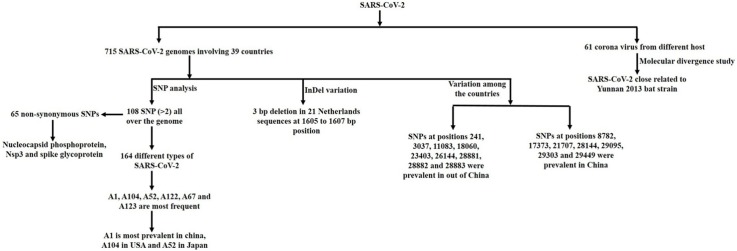
Graphical abstract
Keywords: COVID-19, SARS-CoV-2, SNP, Nucleocapsid phosphoprotein, Nsp3, Spike glycoprotein
Abstract
Coronavirus disease 2019 (COVID-19) is the newly emerging viral disease, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The epidemic sparked in December 2019 at Wuhan city, China that causes a large global outbreak and a major public health catastrophe. Till now, more than 129 million positive cases have been reported in which more than 2.81 million were dead, surveyed by Johns Hopkins University, USA. The diverse symptoms of COVID-19 and an increased number of positive cases throughout the world hypothesize that this virus assembles more variants that are preventing the pursuit of its adequate treatment as well as the development of the vaccine. In this study, 715 SARS-CoV-2 genomes were retrieved from the gisaid and NCBI viral resources involving 39 countries and 164 different types of variants were identified based on 108 Single Nucleotide Polymorphisms (SNPs) in which the ancestral type of SARS-CoV-2 was found as the most frequent and the most prevalent in China. Moreover, variant type A104 was identified as the most frequent in the USA and A52 in Japan. The study also recognized the most common SNPs such as 241, 3037, 8782, 11083, 14408, 23403, and 28144 as well as variants regarding base-pair, C > T. A total of 65 non-synonymous SNPs were recognized which were mostly located in nucleocapsid phosphoprotein, Non-structural protein 3(Nsp3), and spike glycoprotein encoding gene. Molecular divergence analysis revealed that this virus was phylogenetically related to Yunnan 2013 bat strain. This study indicates SARS-CoV-2 frequently alters their genetic material, which mostly affects the nucleocapsid phosphoprotein, and spike glycoprotein-encoding gene and makes it very challenging to develop SARS-Cov-2 vaccine and antibody-mediated rapid diagnostic kit.
1. Introduction
SARS-CoV-2, causing viral infection to humans named as COVID-19 by WHO has rapidly expanded worldwide in an epidemic scale (Zhou et al., 2020; Li et al., 2020). The cluster was first appeared in December 2019 at Wuhan, Hubei Province, China with several symptoms like pneumonia from unknown etiology (Zhu et al., 2020; Xiao et al., 2020; Rothan and Byrareddy, 2020). After Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS), the world is now experiencing the third epidemic as a new public health crisis (Prompetchara et al., 2020). The study showed that the SARS-CoV-2 could be transmitted from person to person by respiratory droplets, fomites, feces, and also through aerosol transmission (Wang et al., 2020a,b; Gu et al., 2020). The etiopathogenesis of COVID-19 is targeting angiotensin-converting enzyme 2 (ACE2) as a viral receptor for initial entry into host cells (Xiao et al., 2020; Abdulamir and Hafidh, 2020) involving multiple pathogenic mechanisms and, infecting epithelial cells of the respiratory tract, later the gastrointestinal tract (Xiao et al., 2020) and so on. Besides, recent studies find evidence of the virus in the cerebrospinal fluid as SARS-CoV-2 can give rise to nervous system damage (Wu et al., 2020a,b,c). In the commencement of the infection, patients either develop mild to moderate symptoms like; fever, cough, fatigue (Wu et al., 2020a,b,c) or severe signs including acute respiratory distress syndrome (ARDS), diarrhea, septic shock, coagulation dysfunction (Giannis et al., 2020), etc. and finally even causes death (Repici et al., 2020). In some cases, patients may stay asymptomatic and cannot be distinguished without the assistance of laboratory tests (Hu et al., 2020; Nishiura et al., 2020).
Typically, coronavirus (CoVs) is a non-segmented and enveloped virus with a positive-sense, single-stranded RNA (Su et al., 2016; Guo et al., 2020). All the virus of Coronaviridae family have crown-like spikes on the outer surface and holds a single strand, positive-sense RNA genome of 26–32 kilobases long, the longest length considering other RNA viruses (Guo et al., 2020). Till now, six coronavirus species are involved in causing human illness (Guo et al., 2020). About, 39 species in 27 subgenera of coronaviruses have been classified where five genera and two subfamilies belong to the family Coronaviridae, suborder Cornidovirineae, order Nidovirales, and realm Riboviria (Abdulamir and Hafidh, 2020; Gorbalenya et al., 2020). Among them, four genera of CoVs found in mammals where the gene source of Alphacoronavirus and Betacoronavirus are similar to bat CoVs (Lau et al., 2013; Monchatre-Leroy et al., 2017). In the 21st century, the outbreaks by SARS coronavirus (SARS-CoV), in 2002 and the MERS coronavirus (MERS-CoV-2), in 2012 consecutively showed ∼79 % and ∼50 % (Abdulamir and Hafidh, 2020; Su et al., 2016) similarity with recent SARS-CoV-2 which was believed to be transmitted initially by the zoonotic reservoir. It has been also estimated that SARS, MERS, and SARS-CoV-2 are primarily transmitted from the bat, which fall into the genus Betacoronavirus subgroups (Wong et al., 2019; Lau et al., 2019). However, the transmission of SARS-CoV-2 from bat is yet not confirmed as CoVs are eminent for their high occurrence of recombination and mutation rates with an average substitution rate of ∼10−4 per year per site, which also allow them to adapt to new hosts and ecological roles (Su et al., 2016; Lau et al., 2013).
The present pandemic due to COVID-19 has surfaced an urgency of developing antiviral drugs or vaccines against the virus. The coronavirus responsible for COVID-19 infection shows a low mortality rate of ∼3-4 % with the highest transmission comparing with SARS-CoV (9% death), MERS-CoV-2 (36 % death) (Su et al., 2016). Looking at the statistics, investigation shows that immune-compromised (Roncon et al., 2020; Fishman and Grossi, 2020) elder individuals are increasingly inclined to see serious conditions due to this infection. The study also forecast that males (Giannis et al., 2020) are significantly more prone to build up the disease rather than females. Increased number of the positive cases creates concerns about the tendency of accumulating more variants in the virus and therefore a widespread genomic variation investigation is required to infer the evolutionary rate, molecular divergence, pathogenesis for developing successful treatment and effective antiviral drug targeting their key enzymes or proteins as well as the vaccine.
In this study, we retrospectively observed genome-wide comparison of 715 SARS-CoV-2 genomes, retrieved from the Global Initiative on Sharing All Influenza Data (GISAID) and National Center for Biotechnology Information (NCBI) viral resources distributed in 39 countries to understand their genomic variation and their origin of evolution with other Coronaviriade strains.
2. Materials and methods
2.1. Genome sequence retrieval
Initially, 1067 SARS-CoV-2 complete genome sequences were retrieved from GISAID (Shu and McCauley, 2017) uploaded before 15 april 2020. Sequences, containing ambiguous letter, especially N with an undetermined number and large gap within the sequences were eliminated and 714 complete SARS-CoV-2 genome sequences (Supplementary Table 1) were selected for further study (Wang et al., 2020a,b). SARS-CoV-2 with GenBank accession no. NC_045512.2 and 52 other viruses from different host belonging to the Coronaviridae family (Supplementary Table 2) were also retrieved from the NCBI viral database.
2.2. Whole genome alignment and variant analysis
The genome sequence of SARS-CoV-2 with GenBank accession no. NC_045512.2 which was collected from Wuhan China in December 2019 and 714 complete SARS-CoV-2 genome sequences were aligned by G-INS-I algorithm with 1000 maxiterate and default parameter using in multiple sequence alignment tool MAFFT version 7.450 (Katoh and Standley, 2013). Then SNP-sites version 2.3.3 with default parameter was used for extraction of single nucleotide polymorphisms (SNPs) from multiple sequence alignment (Page et al., 2016). Insertion Deletion (InDel) variation was called from the vcf format of aligned data.
2.3. Phylogenetic analysis
Two phylogenetic trees were developed in this study, one for understanding the origin of the evolution of SARS-CoV-2 and another to observe their variation among the countries. Eight viral strains from pangolin, isolated from Guangdong and Guangxi of China, one bat strain from Yunnan, China and 52 from different hosts including SARS-CoV and MERS-CoV belong to coronaviridae family were aligned by MAFFT and then MEGA software version 10 with 1000 bootstrap and maximum likelihood method were used for the development of phylogenetic tree (Kumar et al., 2018). Breda virus was used as an out-group. A similar protocol was followed for the development of a country-wise phylogenetic tree except all 715 SARS-CoV-2 sequences were sorted according to their source countries or regions and then the most variable one was selected for representing the respective region.
2.4. Variation analysis at protein coding gene
Polymorphic protein coding sequences were translated by bio biomedicine of the University of Gothenburg translation utility (Sequence bioinformatics-Translation utility, University of Gothenburg, 2020) and aligned with existing protein sequences of SARS-CoV-2 present in NCBI virus database for the identification of synonymous and non-synonymous SNPs.
2.5. Statistical analysis and visualization
All the SARS-CoV-2 genome sequences were grouped in either China or out of China and compared by logistic regression based on SNP that was frequent in at least two sequences. Significant P-values were plotted in mirror Manhattan plot by Hudson R package in R statistics in which all the structural regions of SARS-CoV-2 were ranked in 1–22 including 5′ UTR and 3′ UTR whether Nsp7, Nsp8, Nsp9 and ORF 6, ORF 7a, ORF 7b, ORF 8 were merged and named as 8 and 19 respectively.
3. Results
The retrieved genome sequences of SARS-CoV-2 used in the present study were found to be distributed among 39 countries and all the strains were involved in the current coronavirus outbreak. All the sequences showed almost 99 % sequence similarity even though they acquired a number of several SNPs and InDel variations at different positions, which made them highly variable among the countries even within the countries.
3.1. Identification of Single Nucleotide Polymorphism (SNP)
Because of some sequencing error at both of the tailoring ends, sequences from 35 bp (base pair) to 29705 bp were considered for further analysis. A total 736 SNPs were identified in that (35 bp to 29705 bp) region. Among them 541 were identified in a unique isolates, 87 for two and 108 occurred for more than two isolate at different positions of the genome. SNPs 8782, 28144, 23403, 3037, 241, 14408, 11083, 18060, 17747, 17857, 28881, 28882, 28883, 26144, 1059 and 25563 bp were the most frequent (Fig. 1 ).
Fig. 1.
The genome-wide frequency of 108 SNPs in SARS-CoV-2. SNPs at 241, 3037, 8782, 11083, 14408, 23403, and 28144 were the most frequent and SNPs at 28881, 28882 and 28883 positions represented similar frequency.
3.2. Insertion Deletion (InDel) polymorphismanalyses
InDel variation analysis among the 715 SARS-CoV-2 genomes revealed no significant insertion variation. Seventeen deletion variations were identified at the genomic level (Table 1 ). A 3 bp deletion at 1605–1607 bp position occurred at the highest frequent number 21 and this type of deletion variation only found among the Netherlands SARS-CoV-2 strains. Besides, a 15 bp deletion at 508–523 bp position and a 9 bp deletion at 518–520 bp position were identified for thrice. Among the seventeen deletion variations, five were identified twice and rest of them was found once.
Table 1.
Position wise distribution of deletion variation among the SARS-CoV-2 genome.
| Sl no | Genomic Position (bp) | Deleted sequences | Number of bp | Frequency |
|---|---|---|---|---|
| 1. | 68 | A | 1 | 1 |
| 2. | 239-41 | TTC | 3 | 1 |
| 3. | 253-57 | ACCGA | 5 | 1 |
| 4. | 359-82 | GGAGACTCCGTGGAGGAGGTCTTA | 24 | 1 |
| 5. | 508-23 | TGGTCATGTTATGGT a, b | 15 | 3 |
| 6. | 518-20 | ATG a, c | 3 | 2 |
| 7. | 669-71 | GTT a | 3 | 2 |
| 8. | 686-94 | AAGTCATTT a, d, e | 9 | 3 |
| 9. | 1605-07 | ATGc | 3 | 21 |
| 10. | 6950-57 | ATTATAAT | 8 | 1 |
| 11. | 7222-27 | TGCTTT | 6 | 1 |
| 12. | 11075 | T a, f | 1 | 2 |
| 13. | 11083 | T | 1 | 1 |
| 14. | 12620-22 | TCA c | 3 | 2 |
| 15. | 20301-03 | ATT | 3 | 1 |
| 16. | 21991-93 | TTA c, g | 3 | 2 |
| 17. | 25617-22 | GGTGTT | 6 | 1 |
USA.
Japan.
Netherlands.
Canada.
China Shanghai.
China Anhui.
India.
3.3. Identification of variant type of SARS-CoV-2
By using 108 SNPs, 164 different types of SARS-CoV-2 were identified in which 88 unique types were observed at a single frequency (Fig. 2 ). All the 164 types were named through A1 to A164. The ancestral type (A1) of SARS-CoV-2 which possessed no SNPs was identified as the most frequent for 91 times among the 715 SARS-CoV-2 genomes that was mostly prevalent in China. Besides, A104, A52, A122, A67 and A123 types were identified at 65, 54, 30, 27 and 23 frequencies respectively (Fig. 3 ). A104 and A52 were the most frequent in USA and Japan respectively whether other variants were distributed among the countries.
Fig. 2.
SARS-CoV-2 genetic pattern based on 108 SNPs. 164 different types were identified among the 715 SARS-CoV-2 genomes where the ancestral type (A1) was the most frequent. 88 unique types of SARS-CoV-2 that were observed at a single frequency were excluded from the visualization. The frequencies of the pattern are presented as a color gradient with frequencies is shown on the right. W denotes the wild type.
Fig. 3.
The top most prevalent pattern of SARS-CoV-2. Top 6 frequent patterns with their variable positions are presented here which represent 40.66 percent of the total population.
3.4. Origin of evolution study
To study the origin of the evolution of current outbreak of SARS-CoV-2, 61 other virus belong to Coronaviridae family were retrieved. After phylogeny analysis using the first genome sequenced of SARS-CoV-2 virus (GenBank accession no. NC_045512.2) revealed that current outbreak of SARS-CoV-2 was very close to China Yunnan 2013 bat corona strain (Fig. 4 ). Besides that SARS-CoV-2 was very close with China Guangdong pangolin coronavirus strain which was also identified at the end of 2019.
Fig. 4.
Phylogenetic analysis of current outbreak SARS-CoV-2 with other Coronaviridae viruses. SARS-CoV-2 were grouped with the Yunnan bat 2013 strain and clustered with pangolin strain isolated from Guangdong and Guangxi China where all of them were evolved from ancestral node SARS virus.
3.5. Variation of SARS-CoV-2 among the countries
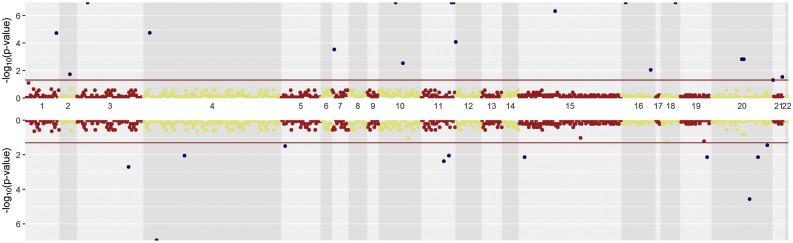
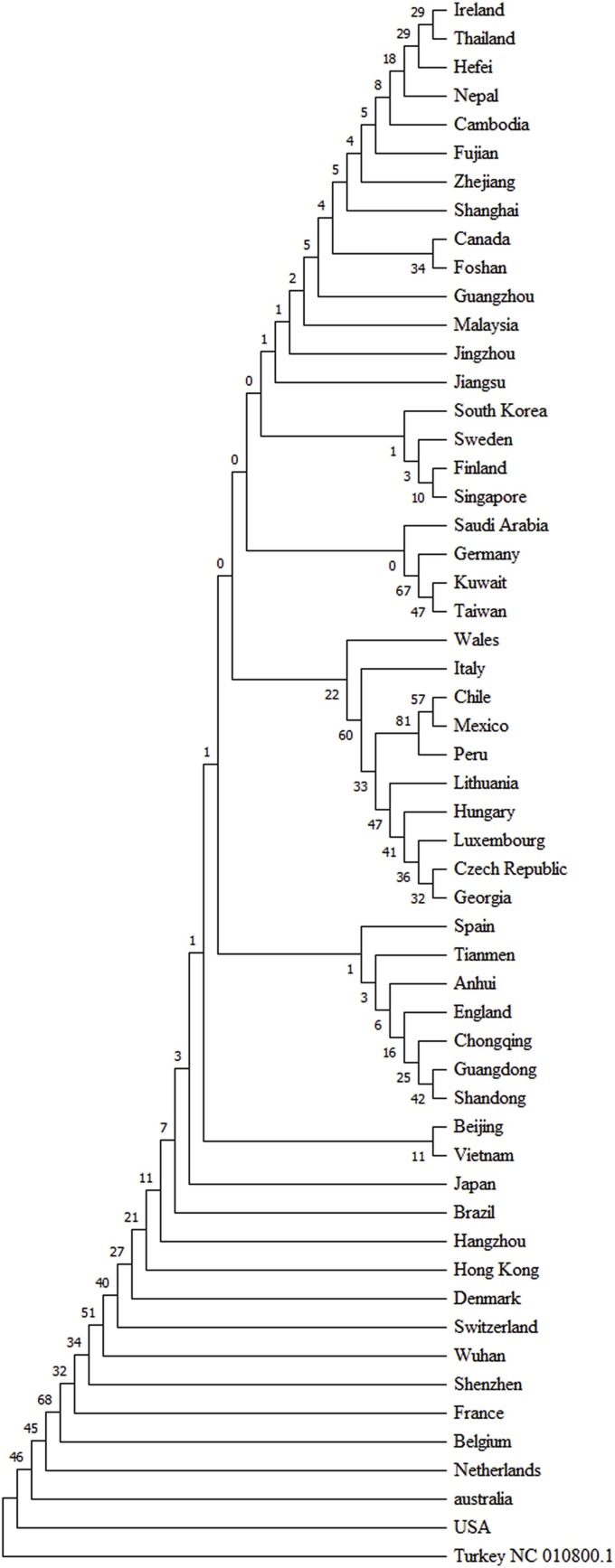
In these study total 715 SARS-CoV-2 genomes were studied which were distributed among the 39 countries. SNPs comparison of SARS-CoV-2 between China and rest of the 38 countries were plotted in Fig. 5 . SNPs at positions 241, 3037, 11083, 18060, 23403, 26144, 28881, 28882 and 28883 were significantly prevalent in other countries than China. On the other hand, SNPs at positions 8782, 17373, 21707, 28144, 29095, 29303 and 29449 were significantly prevalent in China compared to other countries. SNPs at positions 514, 1059, 14408, 14805, 17746, 17858, 25563, 27046, 29553, and 29635 were significantly present only in other countries whether SNPs at 2293, 3171, 3885 and 17502 were only significantly present within China. InDel analysis revealed a 3 bp deletion at 1605 bp position which was present only in the Netherlands at the highest frequency. Besides, different types of deletion variations were mostly present in the Netherlands viral strains at different positions e. g. at 518, 12620 and 21991 positions. On the other hand, 4 deletion variations were identified in USA viral strains at 508, 518, 669 and 686 positions. Deletion variations at 669 and 12620 positions were identified only in USA and the Netherlands respectively whether variation along with countries e. g. at the position of 508 was shared between USA and Japan, position 518 between USA and the Netherlands, position 686 among the USA, Canada and China Shanghai, 11075 between USA and China Anhui and the 21991 position between the Netherlands and India (Table 1). Countries or regionsthat have 5 or more than 5 viral strains were included for the rate of variation analysis. Among them, Japan had the lowest rate of variation and Wales had the highest (Fig. 6 ). Rate of variation in China was 0.44 but some of their provinces such as Shenzhen and Guangzhou had higher and Shandong and Wuhan had lower rate of variation compared to whole China. In USA, it had the second lowest and in Italy it had the second highest rate of variation. Phylogenetic analysis using the viral strains having highest number of polymorphic positions from the 39 countries revealed that most of the countries or regions had distinct variants although some of them shared variant form of SARS-CoV-2 and clustered together (Fig. 7 ).
Fig. 5.
SNP based genome-wide comparison of SARS-CoV-2 viral strain between China and countries other than China. All the structural regions of SARS-CoV-2 were ranked between 1 to 22 including 5′ UTR and 3′ UTR whether Nsp7, Nsp8, Nsp9, and ORF 6, ORF 7a, ORF 7b, ORF 8 were merged and named as 8 and 19 respectively. Upper Manhattan plot represents the significant SNPs out of China and lower represents in China indicated by the deep blue dot. Structural position 20 denotes the nucleocapsid phosphoprotein comprised of with the highest number of significant SNP between China and out of China. Structural positions 5, 6, 8, 9, 13, 14, and 17 representing the Nsp4, 3 C like proteinase, Nsp7+Nsp8+Nsp9, Nsp10, endoRNAse, 2′ -O-ribose methyltransferase, and envelope protein respectively having no significant SNP between China and Out of China.
Fig. 6.
Rate of variation of SARS-CoV-2 among the 18 countries. Japan had the lowest rate of variation; on the other hand, Wales had the highest. In China, Shenzhen provinces had the highest rate of variation and Shandong had the lowest.
Fig. 7.
Phylogenetic analysis of SARS-CoV-2 among the countries or sources of their region. Viral strain having the highest number of the polymorphic regions was selected and it was found that most of the viral strains were distinct from each other even within the countries although some of them were clustered.
3.6. Variation at protein coding gene
Among the 108 SNPs, 100 SNPs were located in the coding region and 35 of them were synonymous SNPs (Supplementary Table 3). SNPs at 4255 bp were changed either by A or T and similarly at the position of 13408 by A or G. Substitution by A at 13408 bp position and SNP at the position 28883 brought stop codon. SNPs at 28881, 28882 and 28883 were changed simultaneously where GGG were substituted by AAT. These three SNPs located in the nucleocapsid phosphoprotein-encoding gene where 28881 and 28882 were at the 2nd and 3rd nucleotide of Arginine amino acid encoding codon at 203 position and 28883 was first nucleotide of Glutamine amino acid encoding codon at 204. After polymorphism, amino acid at 203 position was changed to Lysine followed by a stop codon at 204 position. Consequently, 419 amino acids containing nucleocapsid phosphoprotein shortened to 203. Among the 108 SNPs, none of them was found in Nsp7, Nsp8, Nsp9, 2′ - O -ribose methyltransferase, envelope protein, ORF 7a and ORF 7b encoding gene whereas leader protein, 3 C like proteinase and endoRNAse only had synonymous SNP. Nucleocapsid phosphoprotein, Nsp3 and Spike glycoprotein contained highest number of non-synonymous SNPs in their protein, which were numbered as 16, 9 and 7 respectively.
4. Discussion
SARS-CoV-2, a virus from the Coronaviridae family is responsible for the highly transmissible and pathogenic infection of COVID-19 around the globe (Guo et al., 2020). Until now, multiple epicenters have been identified. Among them, only the Republic of China reported an improved scenario against COVID-19 whereas the condition of Europe and the USA is still deteriorating at an alarming rate. Additionally, countries of Latin America, South Asia, and Eurasia particularly Brazil, India, and Russia respectively have emerged as a new hotspot of COVID-19. Globally the situation has not only startled the community for its rapid community transmission but also created a great concern into the development of the COVID-19 vaccine and efficient treatment option. Therefore, to understand the molecular divergence of SARS-CoV-2, the current study aims to have a genome-wide comparison of the novel coronavirus sequences and their origin of evolution with other Coronaviriade strains.
The previous investigation suggested that SARS-CoV-2 was originated from bat (Zhou et al., 2020; Lu et al., 2020). However, other studies also reported that intermediate mammal e.g. Pangolin may be responsible for the transmission of this virus (Lam et al., 2020). In this study, phylogenetic analysis revealed that SARS-CoV-2, a potential causing agent for COVID-19 was significantly related to Yunnan 2013 bat strain. In addition, the Guangdong 2019 pangolin strain found as a sister group of current corona strain clustered with other Guangxi 2017 pangolin strains. All of these strains were also evolved from SARS.
Since the rate of mutation is high at RNA virus (Tang et al., 2020), we can hypothesize that SARS-CoV-2 can be mutated frequently. In this study, a total of 736 SNPs identified, among them, only 108 SNPs were spotted in more than two sequences. These large numbers of SNPs results made a diverse type of SARS-CoV-2 variant in which only the top 13 variants comprise 52.5 % of the total study population. These large number of SARS-CoV-2 variant indicates their higher rate of variability within a short time. Besides, among these 108 SNPs, only 100 SNPs were found in the protein-coding region in which only 65 of them were nonsynonymous and showed the highest number of variability in nucleocapsid phosphoprotein, Nsp, and spike glycoprotein. Usually, a structural protein that is exposed to the human immune system readily at their very early stage of infection is targeted as an effective antigen for the development of a vaccine (Scarselli et al., 2005; Chaudhuri et al., 2014; María et al., 2017). SARS-CoV-2 comprises a list of structural proteins e.g. spike glycoprotein, membrane protein, envelope protein, nucleocapsid phosphoprotein, etc. (Wu et al., 2020a,b,c). The N terminal domain of the nucleocapsid phosphoprotein (N protein) capture the corona virus genome while C terminal domain anchors the viral membrane through membrane glycoprotein interaction and regulates the viral life cycle (Lu et al., 2021). While the Spike protein (S protein) interacts with the host ACE2 receptor and mediates the membrane fusion (Lu et al., 2021). As a consequences, N protein and S protein would be a potential target for antiviral drug and several compound have been predicted against them (Hu et al., 2021). Moreover, the spike protein in corona virus regarded as a potential antigen and elicited the host immunity by activating the CD4+ helper T cell and CD8+ killer T cell (Grifoni et al., 2020). The study also identified that except envelope protein all of the structural protein contains several numbers of non-synonymous SNPs, which make the virus very tenacious against the development of an effective vaccine as well as the therapeutic inhibitors to SARS-CoV-2 treatment.
The previous study reported that three in-frame deletions took place in the leader protein of ORF1ab and all the non-coding deletions happened in either 5′ UTR or 3′ UTR (Koyama et al., 2020). No significant insertion variations reported by Koyama et al. (Koyama et al., 2020) similarly observed in the current study. Additionally, both in-frame and frameshift deletions were observed all over the genome. Similar to Koyama et al. (Koyama et al., 2020), the study also reported C to T as the most variant frequent in 42 distinct positions where the synonymous SNP found at 8782 bp position and the non-synonymous SNP at 28144 bp position which were most frequent in 204 and 201 respectively in distinct viral strains with the highest prevalence in China. On the other hand, the analysis also showed a significant frequency of the ancestral type (A1) of SARS-CoV-2, which was highest in the Chinese population (34.6 %) among the 164 types whereas the second-highest frequency was observed in the USA (10.4 %). Conversely, the second most prevalent SARS-CoV-2 type (A104) was also found in the USA population with greater frequency (36.6 %) and third prevalent SARS-CoV-2 type, A52 in the Japanese population (62.1 %).
5. Conclusion
This recent pandemic has highly affected our daily lives including public health, mental health, medical, economic growth, education, and so on. This is very evident that SARS-CoV-2 can change their genetic material very frequently and not one type can be conserved among the geographic location. Higher number of the distinct polymorphic regions in structural protein makes this virus very tenacious to rapid diagnosis by antibody, to treat the virus infected patients by antiviral drug targeting spike protein, envelope protein, RNA dependant RNA polymerase etc. and to control the viral prevalence by vaccine mediated development of immunity. Though few vaccines were approved by different countries, current vaccines were not so much trustworthy to fight against the new variant of SARS-CoV-2. Therefore, the present findings on the genomic variations of SARS-CoV-2 would provide insight for further researches on the development of an effective vaccine or treatment.
Financial supports
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Availability of data and materials
All the dataset used in this study were retrieved from public repository of NCBI and GISAID and their accession IDs are mentioned in supplementary documentsand all the softwareused are mentioned in materials and methods section.
CRediT authorship contribution statement
Avizit Das: Conceptualization, methodology, software, data curation, formal analysis, visualization, writing-original draft and editing. Sarah Khurshid: Conceptualization, data acquisition, visualization and review. Aleya Ferdausi: Data acquisition, visualization, review and editing. Eshita Sadhak Nipu: Conceptualization, data acquisition, interpretation and review. Amit Das: Conceptualization, data acquisition, visualization and review. Fee Faysal Ahmed: Data visualization and review.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
None.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.compbiolchem.2021.107533.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Abdulamir A.S., Hafidh R.R. The possible immunological pathways for the variable immunopathogenesis of COVID-19 infections among healthy adults, elderly and children. Electron. J. Gen. Med. 2020;17(4):em202. doi: 10.29333/ejgm/7850. [DOI] [Google Scholar]
- Chaudhuri R., Kulshreshtha D., Raghunandanan M.V. Integrative immunoinformatics for Mycobacterial diseases in R platform. Syst. Synth. Biol. 2014;8(1):27–39. doi: 10.1007/s11693-014-9135-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman J.A., Grossi P.A. Novel Coronavirus-19 (COVID-19) in the immunocompromised transplant recipient: #Flatteningthecurve. Am. J. Transplant. 2020:1–3. doi: 10.1111/ajt.15890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannis D., Ziogas I.A., Gianni P. Coagulation disorders in coronavirus infected patients: COVID-19, SARS- CoV-1, MERS-CoV and lessons from the past. J. Clin. Virol. 2020;127 doi: 10.1016/j.jcv.2020.104362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R.S. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J., Han B., Wang J. COVID-19: gastrointestinal manifestations and potential fecal-oral transmission. Gastroenterology. 2020;158(6):1518–1519. doi: 10.1053/j.gastro.2020.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.R., Cao Q.D., Hong Z.S. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak – an update on the status. Mil. Med. Res. 2020;7(11):1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Song C., Xu C. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci. China Life Sci. 2020;63(5):706–711. doi: 10.1007/s11427-020-1661-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Zhou Z., Li F. The study of antiviral drugs targeting SARS-CoV-2 nucleocapsid and spike proteins through large-scale compound repurposing. Heliyon. 2021;7(3) doi: 10.1016/j.heliyon.2021.e06387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Institute of Biomedicine. July 1, 2020. The Sahlgrenska Academy, University of Gothenburg. http://bio.lundberg.gu.se/edu/translat.html.
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Platt D., Parida L. Variant analysis of COVID-19 genomes. Bull. World Health Organ. 2020;98(7):495–504. doi: 10.2471/BLT.20.253591. 32742035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.T.-Y., Shum M.H.-H., Zhu H.-C. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020:1–6. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- Lau S.K.P., Li K.S.M., Tsang A.K.L. Genetic characterization of betacoronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of pipistrellus bat coronavirus HKU5 in Japanese pipistrelle: implications for the origin of the novel Middle East respiratory syndrome coronavirus. J. Virol. 2013;87(15):8638–8650. doi: 10.1128/JVI.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K.P., Luk H.K.H., Wong A.C.P. Identification of a novel betacoronavirus (merbecovirus) in amur hedgehogs from China. Viruses. 2019;11(11):980. doi: 10.3390/v11110980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Geng M., Peng Y. Molecular immune pathogenesis and diagnosis of COVID-19. J. Pharm. Anal. 2020;10(2):102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Ye Q., Singh D. The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. Nat. Commun. 2021;12(502) doi: 10.1038/s41467-020-20768-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- María R.R., Arturo C.J., Alicia J.A. In: Vaccines. Afrin F., Hemeg H., Ozbak H., editors. IntechOpen; Croatia, Rijeka: 2017. The impact of bioinformatics on vaccine design and development; pp. 123–145. [DOI] [Google Scholar]
- Monchatre-Leroy E., Boué F., Boucher J.M. Identification of alpha and beta coronavirus in wildlife species in france: bats, rodents, rabbits, and hedgehogs. Viruses. 2017;9(12):364. doi: 10.3390/v9120364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiura H., Kobayashi T., Miyama T. Estimation of the asymptomatic ratio of novel coronavirus infections (COVID-19) Int. J. Infect. Dis. 2020;94:154–155. doi: 10.1016/j.ijid.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page A.J., Taylor B., Delaney A.J. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genom. 2016;2(4) doi: 10.1099/mgen.0.000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020;38:1–9. doi: 10.12932/AP-200220-0772. n.d. [DOI] [PubMed] [Google Scholar]
- Repici A., Maselli R., Colombo M., et al. Coronavirus (COVID-19) outbreak : what the department of endoscopy should know. Gastrointest. Endosc. 2020;92(1):192–197. doi: 10.1016/j.gie.2020.03.019. 32179106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncon L., Zuin M., Rigatelli G. Diabetic patients with COVID-19 infection are at higher risk of ICU admission and poor short-term outcome. J. Clin. Virol. 2020;127 doi: 10.1016/j.jcv.2020.104354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020;109 doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarselli M., Giuliani M.M., Adu-Bobie J. The impact of genomics on vaccine design. Trends Biotechnol. 2005;23(2):84–91. doi: 10.1016/j.tibtech.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data – from vision to reality. EuroSurveillance. 2017;22(13) doi: 10.2807/1560-7917.ES.2017.22.13.30494. PMCID: PMC5388101. http://gisaid.org. July 1, 2020. Hosted by the Federal Republic of Germany, 2008-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016;24(6):490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Wu C., Li X., et al. On the origin and continuing evolution of SARS-CoV-2. Nat. Sci. Rev. 2020;7(6):1012–1023. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang Y., Chen Y. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID‐19) implicate special control measures. J. Med. Virol. 2020;92(6):568–576. doi: 10.1002/jmv.25748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Liu Z., Chen Z. The establishment of reference sequence for SARS ‐ CoV ‐ 2 and variation analysis. J. Med. Virol. 2020;92(6):667–674. doi: 10.1002/jmv.25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A.C.P., Li X., Lau S.K.P. Global epidemiology of bat coronaviruses. Viruses. 2019;11(2):174. doi: 10.3390/v11020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Xu X., Chen Z. Nervous system involvement after infection with COVID-19 and other coronaviruses. Brain Behav. Immun. 2020;87:18–22. doi: 10.1016/j.bbi.2020.03.031. 32240762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.C., Chen C.S., Chan Y.J. The outbreak of COVID-19: An overview. J. Chin. Med. Assoc. 2020;83(3):217–220. doi: 10.1097/JCMA.0000000000000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Tang M., Zheng X. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158(6):1831–18333. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the dataset used in this study were retrieved from public repository of NCBI and GISAID and their accession IDs are mentioned in supplementary documentsand all the softwareused are mentioned in materials and methods section.