Abstract
The acute respiratory syndrome COVID-19 disease, which is caused by SARS-CoV-2, has infected many people over a short time and caused the death of more than 2 million people. The gold standard in detecting COVID-19 is to apply the reverse transcription polymerase chain reaction (RT-PCR) test. This test has low sensitivity and produces false results of approximately 15%–20%. Computer tomography (CT) images were checked as a result of suspicious RT-PCR tests. If the virus is not infected in the lung, the virus is not observed on CT lung images. To overcome this problem, we propose a 25-depth convolutional neural network (CNN) model that uses scattergram images, which we call Scat-NET. Scattergram images are frequently used to reveal the numbers of neutrophils, eosinophils, basophils, lymphocytes and monocytes, which are measurements used in evaluating disease symptoms, and the relationships between them. To the best of our knowledge, using the CNN together with scattergram images in the detection of COVID-19 is the first study on this subject. Scattergram images obtained from 335 patients in total were classified using the Scat-NET architecture. The overall accuracy was 92.4%. The most striking finding in the results obtained was that COVID-19 patients with negative RT-PCR tests but positive CT test results were positive. As a result, we emphasize that the Scat-NET model will be an alternative to CT scans and could be applied as a secondary test for patients with negative RT-PCR tests.
Keywords: Scattergram, Reverse transcription polymerase chain reaction, Computer tomography, Convolutional neural network
1. Introduction
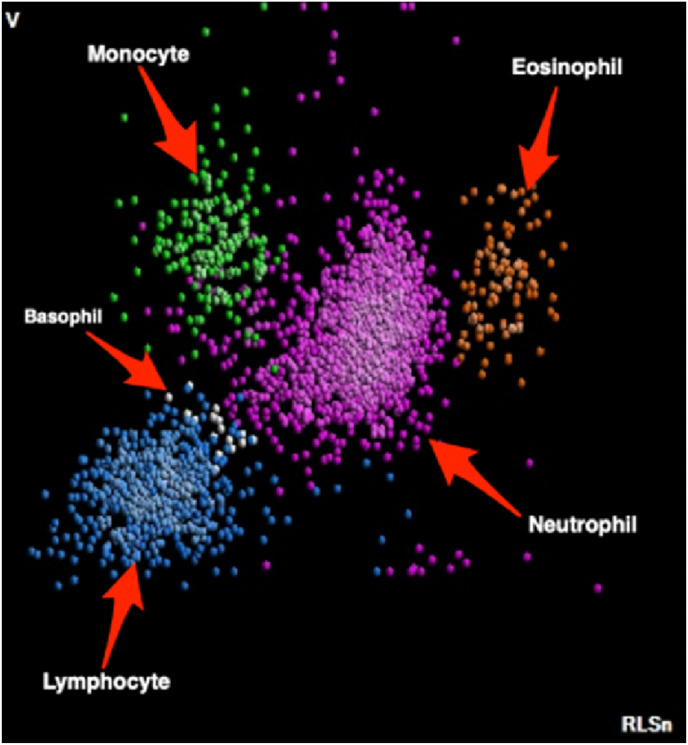
Complete blood count (CBC) is one of the routine tests used frequently during the initial evaluation of patients. CBC analysers such as DXH-800 (Beckman Coulter) provide a differential fluorescence scattering diagram (WBC differential fluorescence - WDF) that displays a classification of white blood cells (WBCs) according to their morphology and intracellular components. Each type of leukocyte is always displayed in the same area, and different types have different colours. The different leukocyte clusters displayed on the WDF scatter diagram are matched with optical microscopy and visual inspection and provide the user with the scattergram image in Fig. 1 .
Fig. 1.
CBC-DIFF scattergram image.
Automated haematology analysers are often used in clinical laboratories to assess the health status of patients. These analysers provide detailed information about WBCs. WBCs are a type of immune cell that helps fight infections and other diseases. WBCs or leukocytes are important elements of the immune system that help the body fight infections. The different types of WBCs are basophils, neutrophils, eosinophils, lymphocytes, and monocytes. Relationships can be established between WBCs found in red blood cells in both men and women and weight, uric acid, cholesterol, heart rate, creatinine, ethnicity, sex, height, blood pressure, and blood sugar. Abnormalities in the distribution of cells on the scattergram are important indicators in determining many other diseases, such as leukaemia [[1], [2], [3]].
Recent reports have shown that some COVID-19 patients have high leukocyte counts [4,5]. Clinical data on these patients are important. Indeed, many researchers have examined the relationship between severe courses of COVID-19 and white blood cell counts in the body [6,7]. In these studies, it was revealed that some patients with COVID-19 had increased WBC and neutrophil counts with a decrease in lymphocyte count. In addition, the neutrophil-lymphocyte ratio has been shown to be a biomarker that can predict the outcome of infection (https://www.news-medical.net/news/20.201.019/The-connection-between-severe-COVID-19-and-white-blood-cell-counts.aspx).
In a pioneering study that considered whether the white cell distribution could be an indicator of COVID-19, Osman et al. emphasized that the presence of plasmacytoid lymphocytes in the white blood cell distribution diagram indicates suspicion of COVID-19. The findings showed that COVID-19 patients had a sensitivity of 85.9% and a specificity of 83.5% [8]. Mitra et al. observed that CBC can be associated with COVID-19 and observed changes in neutrophil cells. A 46-year-old, previously healthy woman had flu-like symptoms, and chest X-ray and chest CT showed signs of lobar pneumonia. Subsequent radiology studies found that lung findings worsened and that the patient (COVID-19) was positive. Leukocytosis (14.1 × 103/μL) and peripheral blood smears were observed with the patient's left-shifted neutrophilic cell population after the first complete blood count [9]. Anurag et al. studied the relationship between COVID-19's different haematological parameters and the disease severity. They analysed the effects of hypertension and diabetes on the neutrophil to the lymphocyte ratio and neutrophil-to-monocyte ratio in patients suffering from COVID-19. The study consisted of data on 148 SARS-CoV-2 infections confirmed by physicians. As a result, they stated that leukocytes, neutrophils, lymphocytes, monocytes and eosinophils, a high neutrophil-monocyte ratio (NMR) and a neutrophil-lymphocyte ratio (NLR), are indicators of disease severity in COVID-19 [6]. Zhao et al. evaluated the clinical findings of COVID-19 patients with increased leukocytes. Group t tests were used for the evaluation of normally distributed variables, and Mann-Whitney U tests were used for non-normally distributed continuous variables. As a result of the study, it was determined that the number of leukocytes increased and that there was a high number of neutrophils in the peripheral blood in patients with COVID-19 [10]. Sun et al. investigated whether white blood cell characteristics could have potential effects on severe COVID-19. Using Mendelian randomization, an increased percentage of eosinophils of WBCs, myeloid white blood cell count, and granulocyte count have been associated with a high risk of severe COVID-19 [11]. Zhao et al. aimed to analyse the clinical characteristics and abnormal immunity of those who died from COVID-19. A total of 125 patients who died and a total of 414 randomly selected patients with COVID-19 were used as controls. The results were evaluated using the logistic regression algorithm. According to the results obtained, neutrophils, lymphocytes, low CD4+ T cells and decreased C3 are immune-related risk factors that predict the mortality of COVID-19 patients [12]. Selim et al. stated that the leukocyte and lymphocyte counts in COVID-19 patients are reliable indicators of SARS CoV-2 infection [5]. Martens et al. showed that there was a significant decrease in the numbers of leukocytes, platelets and absolute neutrophils in COVID-19 patients. In addition, they stated that lymphocyte forward scattering and reactive lymphocytes were high [13].
1.1. Motivation
RT-PCR is considered to be the basic standard diagnostic search for SARS-CoV-2 that causes COVID-19. However, RT-PCR can produce false results between 15% and 20%. In cases that the doctor deems to be suspicious, the patient's CT images are checked. If the COVID-19 virus is not infected in the lung, this test result is also stated as negative. In this paper, we propose a CNN-based model that can be used in place of CT scans and eliminate suspicious situations in RT-PCR and CT scans. The proposed method makes the diagnosis of COVID-19 accurate, faster and reliable. The proposed system uses scattergram images obtained from the WBC blood test taken from the patients. The scattergram shows the distribution of white blood cell types (basophils, neutrophils, eosinophils, lymphocytes and monocytes) in the 2D plane. Scattergram images obtained from patient and non-patient subjects determined by RT-PCR and CT images are first classified with pretrained CNN (Alexnet, Googlenet and Resnet18) architectures and then with the Scat-NET architecture. With the results obtained, we claim that Scat-NET will be a complementary test to RT-PCR and CT scans and that COVID-19 can be diagnosed from scattergram images.
1.2. Novelties and contributions
In this paper, a 25-depth CNN model was used to detect COVID-19 from scattergram images. The novelties are the following:
-
-
For the first time, COVID-19 detection was conducted with scattergram images.
-
-
Cell numbers such as basophils, neutrophils, eosinophils, lymphocytes and monocytes and the ratios between them are used in the diagnosis of the disease. With this study, all of the parameters were evaluated together, and it was shown that scattergram images obtained from the parameters could be used in the diagnosis of COVID-19.
The contributions of this study are the following:
Although hand-crafted feature extraction is effective, it is not effective in complex computer vision problems. On the other hand, CNN models can successfully extract features. Features extracted from scattergram images were classified with the 25-depth Scat-NET model, and COVID-19 was identified.
-
-
In suspicious cases with negative RT-PCR test results, a less costly model that uses scattergram images equivalent to CT scans was proposed instead of CT scans.
-
-
With this basic and effective method, cases that cannot be detected by CT scans can be detected.
-
-
The proposed model was developed to assist medical doctors in the diagnosis of COVID-19 and to be an alternative test.
1.3. Paper outline
The remainder of the paper is organized as follows. Section 2 presents the related studies. Section 3 introduces the data and the properties to be used for the proposed method. Section 4 explains the proposed system, Section 5 presents the experimental results, and Section 6 gives the discussion. Finally, in Section 7, the conclusions and future work are discussed.
2. Related work
In this section, we describe some remarkable studies presented in the literature on the detection of COVID-19 from RT-PCR tests and CT images, which directly affect the development of this study.
The virus named SARS-CoV-2, the new coronavirus responsible for COVID‐19, has brought the health community and the whole world to face great difficulties and increases this difficulty with its new mutations. Early diagnosis of COVID‐ 19 and controlling the effect of infected patients on viral spread are extremely important in pandemic management. The standard test for COVID‐19 is the RT-PCR test used to detect viral RNA (ribonucleic acid) from clinical samples. The disadvantages of RT-PCR are waiting for 3–4 h to obtain the test results and having false negative rates as high as 15–20%. In addition, there is a need for certified laboratories, trained personnel and expensive equipment. The World Health Organization recommends a second RT-PCR test in suspicious cases where the RT-PCR test is negative. However, studies show that the second test is less effective. Therefore, in the detection of COVID-19, lung CT images are examined by the doctor as a result of the suspicious RT-PCR test. However, COVID-19 might not be detected in CT image checks. The reason is that the virus has not yet infected the lungs. Therefore, the incompatibility between RT-PCR test results and CT findings is confusing for specialist doctors in determining COVID-19. To overcome this problem, machine learning-based decision support systems are being developed to assist doctors.
Both machine learning and statistical methods have been developed to evaluate the diagnostic value of CT and real-time reverse transcriptase-polymerase reaction (rRT-PCR) used in the diagnosis of COVID-19 disease. Long et al. determined the sensitivity of CT and rRT-PCR tests by including all patients with suspected COVID-19 and pneumonia who were examined by both CT and RT-PCR at first admission. They suggested repeating the rRT-PCR test in patients with positive CT findings because rRT-PCR could give false negative results [14]. Trisnawati et al. stated that multiple RT-PCR tests were performed 11, 8, 11 and 14 times in 4 different patients, and both negative and positive results were obtained [15]. Brinati et al. studied 279 patients with the rRT-PCR test. A total of 177 of these patients were positive, and 102 were negative. They proposed a machine learning model to distinguish patients who are positive or negative for SARS-CoV-2. According to the results, the accuracy was 86%, and the sensitivity was 95%. In this study, a decision support system was developed to interpret blood tests for suspected COVID-19 cases determined by the rRT-PCR test. As a result, the feasibility and clinical robustness of using blood test analysis and machine learning as an alternative to rRT-PCR to identify COVID-19-positive patients have been demonstrated [16]. Cabitza et al. proposed a system that uses machine learning algorithms as an alternative to RT-PCR testing. They analysed the results with routine blood tests, such as haematological and coagulation tests. The AUC was 0.75–0.78, and the specificity was 0.92–0.96 [17].
A number of deep learning-based methodologies have recently been proposed to automate the detection of COVID-19 from CT scans and to assist doctors. In addition to the known deep learning models in these studies [[18], [19], [20]], ensemble deep learning [21], multitask deep learning [22], weakly supervised deep active learning [23], and bidirectional long short-term memory [24] models have also been used.
Ardakani et al. proposed a decision support system for the diagnosis of COVID-19 from CT imagery using 10 different deep learning techniques. The AUC value with Xception and ResNet-101, where the best performance was obtained, was 0.994 (specificity, 99.02%; accuracy, 99.51%; sensitivity, 100%), and 0.994 (specificity, 100%; accuracy, 99%; sensitivity, 98.04%). As a result, they suggested that the decision support system as a high-precision model can be used as an auxiliary tool in radiology departments [25]. Khan et al. proposed a deep learning framework to distinguish COVID-19 from pneumonia. In the proposed model, an extreme learning machine classifier was used, together with a 15-layer CNN that extracts deep features. The average accuracy, sensitivity, specificity and precision values were 95.1%, 95.1%, 95% and 94%, respectively [26]. Amyar et al. proposed a multi-task deep learning architecture to both detect and classify COVID-19 lesions in CT scans. Dice and accuracy were achieved at 88% and 94.67%, respectively [22]. Aslan et al. classified COVID-19 with 98.70% accuracy with a hybrid model based on bidirectional long short-term memory. Although COVID-19 is diagnosed with high accuracy in the aforementioned studies, there are cases where COVID-19 could not be detected in either RT-PCR or CT scans. The underlying reason for this finding is that RT-PCR has a false negative rate of approximately 15–20% [24].
The most important disadvantage in the diagnosis of COVID-19 from CT images is that the result is negative if the COVID-19 virus has not yet infected the lungs. Complementary studies and methods are needed to eliminate the shortcomings of both RT-PCR and CT tests and to increase the sensitivity of the first test of COVID‐19 patients. Therefore, in this article, a CNN-based Scat-NET model was developed to determine COVID-19 from scattergram images obtained from WBC tests.
3. Data
In this study, a total of 335 patients with suspected COVID-19 who did not have any chronic diseases before entering Elazig City Hospital between 01.10.2020 and 01.01.2021 were asked for RT-PCR, CBC and CT scanning at the time of application. A total of 135 of 335 patients were determined to be COVID-19 patients. The PCR test of 121 of these patients was positive, and the PCR tests of other patients were recorded as negative. CT scans of 14 patients with negative PCR tests were performed, and these patients were identified as positive. COVID-19 findings were not found as a result of CT scans in 50 of 135 patients. Both RT-PCR and CT scans of the remaining 200 patients were negative. For each patient, CBC-DIFF(Complete Blood Count with Differential) scattergrams and WBC subgroup values (Neu, Lym, Mon, Bazo, Eos) were obtained with a DXH-800 (Beckman Coulter, Inc., Miami, FL, USA) device. The DXH-800 analyser uses the volume, conductivity, and scatter parameters for controlled flow cytometric analysis of WBC differential detection. In addition, other laboratory data of the patients were controlled by CT scans, CRP, D-Dim and ferritin values, and RT-PCR (±) test results were recorded. A few examples of the data used are given in Fig. 2 . Fig. 2a and 2.b shows scattergram images with positive and negative COVID-19 diagnoses, respectively.
Fig. 2.
COVID-19 and non-COVID-19 scattergram images.
4. The proposed system
As mentioned earlier in this article, we focus on developing a CNN-based model to detect COVID-19 from scattergram images. We chose a model that could lead us to obtain the best benefit in terms of the classification performance.
If the RT-PCR test was negative, CT scans were requested from suspected patients. In addition to the cost of CT scans, erroneous results can be obtained if the virus has not yet infected the lungs. To avoid this disadvantageous situation, we propose a Scat-NET model that can diagnose COVID-19 from scattergram images. The model is a 25-depth CNN architecture for the classification of scattergram images, which can be easily obtained from WBC tests requested from almost every patient. The structure of the proposed model is shown in Fig. 3 .
Fig. 3.
The proposed method.
CNN is the most commonly used model among deep learning models. This architecture has been widely used in medical image processing applications, especially in recent years. A CNN is created by using one or more of the convolutional, activation function, pooling, dropout, and fully connected layers. CNNs are usually designed to follow each other and provide feature models from deep-level features to high-level features of images. In our proposed model, there are 5 layers of convolution, normalization, ReLU and pooling. Additionally, to classify the feature vectors in the last pooling, the CNN contains fully connected, softmax and classification layers. The process of determining COVID-19 with Scat-NET is as follows:
-
Step 1
: Scattergram images containing COVID-19 (−) and COVID-19 (+) cases obtained from the WBC blood test were obtained.
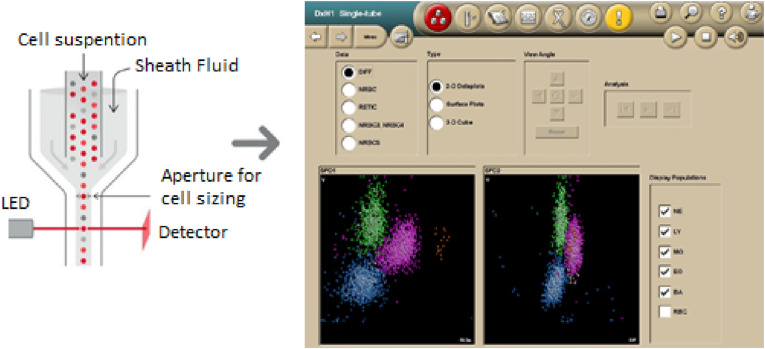
Scattergram images are obtained from patients. A DXH-800 device was used to obtain neutrophils, eosinophils, lymphocytes and monocytes from WBCs in the 2D plane. This device includes a light emitting diode (LED) or laser, as shown in Fig. 4 . With the use of LEDs, light cells pass through; as a result, the light is perceived at a zero angle. As the scattered light passes through the cells, the cell structure is reflected by the absorbed light. Changes in impedance indicate the cell size. The representation of the obtained cell dimensions in the 2D plane is called a scattergram. Scattergram images obtained from patient WBC samples constitute the input of the proposed system. Fig. 4 shows the scattergram image obtained from the WBC blood sample.
-
Step 2
: Give data as an input to the model and train the proposed model. In CNN models, a set of convolution, normalization, ReLu and pooling layers are applied on each image. Due to these steps, the feature vectors of each image are obtained. The classification process is completed by applying dropout, fully connected and softmax operations to the feature vectors. The basic layer properties of the Scat-NET architecture are given in Fig. 5.
Fig. 4.
Obtaining the scattergram images.
Fig. 5.
Layer properties of the Scat-NET.
Convolution Layer: This layer is the main building block of the CNN. This layer, which is responsible for perceiving the properties of the picture, is the most important component of the CNN. This layer applies some filters to the image to extract low- and high-level features in the image. Filters can be of different sizes, such as 3 × 3, 2 × 2. Due to the filters, the output data are created by applying the convolution process to the images from the previous layer. As a result of the convolution process, an activation map of the image is formed. The activation map gives the regions where features specific to each filter are discovered. In other words, it determines which parts of the image are important.
Normalization: In this layer, which is used to reduce the CNN training time, each element is normalized by the expression in Eq. (1). The normalization process is performed using a certain number of neighbouring channels. In Eq. (1), x’ is the normalized value, and x is the input element.
| (1) |
K, α, and β are the hyperparameters in the normalization, and ss is the sum of squares of the elements in the normalization window.
ReLU: ReLU is the most commonly used activation function in models developed based on deep neural networks. One of the most important features of this layer is that it pulls the negative values in the input data to zero. Thus, faster learning of the network is provided. The ReLu function used after the convolution and normalization layers is given in Eq. (2).
| (2) |
Pooling: The purpose of using the pooling layer after the ReLU layer is to reduce the number of parameters and dimensions of the network. There are two types of pooling methods. The first is the average pooling method, which takes the average of the values in the pixels the filter is applied to, and the second is the maximum pooling method, which takes the maximum value in the pixels. The maximum pooling method is often used because of its better performance.
Dropout: The dropout layer is used during training to eliminate some of the neurons to prevent overfitting. If the network enters an excessive learning process, it loses its ability to learn. The drop layer randomly sets the input items to zero with a given probability. Thus, the network is prevented from memorizing.
Fully Connected: The fully connected layer (FC) operates on an input where each input is connected to all neurons. FC layers are often found towards the end of the CNN architecture and are used to optimize goals such as class scores.
Softmax and Classification Layers: For classification problems, a softmax layer followed by a classification layer follows the last fully connected layer. The output unit activation function is given in Eq. (3).
| (3) |
where 0 ≤ yr ≤ 1 and.
-
Step 3
: Train the Scat-NET model based on optimized hyperparameters. The hyperparameters used for training the network are shown in Table 1 .
-
Step 4
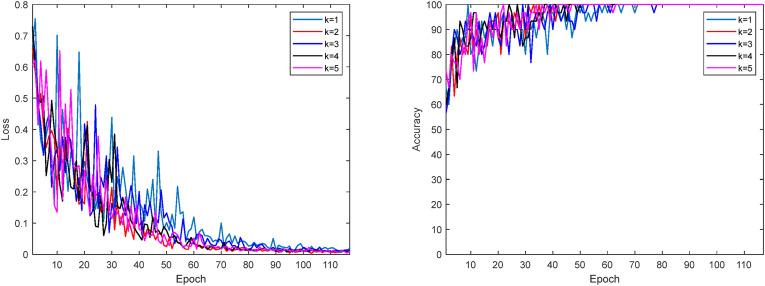
: Apply the k-fold validation model to prevent over-fitting. Cross-validation is applied to fix the overfitting problem to provide a check on the errors. Thus, healthier classification results are obtained, and it is checked whether the classification has been made correctly. Fig. 6 shows the change in the loss function and accuracy for each value of k. The loss function is calculated as given in Eq. (4). K indicates the number of classes, and N indicates the number of samples. Here, t ij indicates that the ith instance belongs to the jth class, and y ij denotes the output of the ith instance for the jth class.
| (4) |
-
Step 5
: Evaluation of the results. The proposed model uses a system with 8 GB RAM, an i7 9750H processor, and a GeForce GTX 1050 PC and was implemented with the MATLAB package program. The performance parameters are the sensitivity (S), specificity (Sp), F1-score, Matthews correlation coefficient (M), area under curve (AUC) and accuracy (A), calculated from the confusion matrix obtained in the experimental results. True positive (TP), false positive (FP), true negative (TN) and false negative (FN) values were used to calculate the measurements. The metrics calculated with these values are given in Table 2 .
Table 1.
Hyperparameters used in the training process of the Scat-NET.
| Parameter | Value |
|---|---|
| Initial Learn Rate | 0.001 |
| Execution Environment | Cpu |
| Max Epoch | 15 |
| Validation Frequency | 3 |
| Mini Batch Size | 3 |
| Optimizer | Stochastic Gradient Descent (SGDM) |
Fig. 6.
Loss function and accuracy change.
Table 2.
Performance parameters.
| S = TP/(TP + FN) |
|---|
| SP = TN/(FP + TN) |
| P = TP/(TP + FP) |
| NPV = TN/(TN + FN) |
| FPR = FP/(FP + TN) |
| FDR = FP/(FP + TP) |
| FNR = FN/(FN + TP) |
| A = (TP + TN)/(P + N) |
| F1 = 2TP/(2 TP + FP + FN) |
| M = TP*TN-FP*FN/sqrt((TP + FP)*(TP + FN)*(TN + FP)*(TN + FN)) |
| AUC = 1/2 ((TP/(TP + FN))+(TN/(TN + FP))) |
S: sensitivity, SP: specificity, P: precision, NPV: negative predictive value.
FPR: false positive rate, FDR: false discovery rate, FNR: false negative rate.
A: accuracy, F1: F1 Score, M: Matthews correlation coefficient, AUC: area under curve.
5. Experimental results
In this section, we present the performance of alexnet, googlenet, Resnet18 and Scat-NET in classifying scattergram images.
In experimental studies, 30% of the 335 scattergram images were used for testing, 10% for validation and 60% for training. First, the Alexnet, Googlenet and Resnet18 models were used to classify COVID-19 cases using scattergram images. The performance parameters obtained are shown in Table 3 .
Table 3.
Pretrained model performance parameters.
| S | SP | P | NPV | FPR | FDR | FNR | A | F1 | M | AUC | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Alexnet | 89.47 | 90.32 | 85 | 93.33 | 9.68 | 15 | 10.53 | 90 | 87.18 | 79.06 | 89.89 |
| Googlenet | 86.84 | 88.71 | 82.5 | 91.67 | 11.29 | 17.50 | 13.16 | 88 | 84.62 | 74.86 | 87.77 |
| Resnet18 | 85.37 | 91.53 | 87.50 | 90 | 8.47 | 12.5 | 14.63 | 89 | 86.42 | 77.20 | 88.45 |
The classification success of the proposed Scat-NET was determined by applying k-cross validation to the data. Cross validation was conducted 5 times. The performance parameters and their average values are given in Table 4 . Confusion matrices for each iteration are given in Fig. 7 .
Table 4.
Performance parameters.
| The proposed method performance parameters (%) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Iteration | S | SP | P | NPV | FPR | FDR | FNR | A | F1 | M | AUC |
| 1 | 92.31 | 93.44 | 90 | 95 | 6.56 | 10 | 7.69 | 93 | 91.14 | 85.37 | 92.88 |
| 2 | 91.89 | 90.48 | 85 | 95 | 9.52 | 15 | 8.11 | 91 | 88.31 | 81.18 | 91.18 |
| 3 | 94.74 | 93.55 | 90 | 96.67 | 6.45 | 10 | 5.26 | 94 | 92.31 | 87.47 | 94.15 |
| 4 | 91.89 | 90.48 | 85 | 95 | 9.52 | 15 | 8.11 | 91 | 88.31 | 81.18 | 91.18 |
| 5 | 90.48 | 96.55 | 95 | 93.33 | 3.45 | 5 | 9.52 | 94 | 92.68 | 87.68 | 93.52 |
| Average | 92.26 | 92.9 | 89 | 95 | 7.1 | 11 | 7.74 | 92.4 | 90.55 | 84.58 | 92.58 |
Fig. 7.
Confusion matrices obtained by applying k-cross validation.
6. Discussion
The female (age mean = 41.25) and male (age mean = 42.62) numbers of 135 COVID-19 patients (age mean ± SD = 41,9 ± 10,6) were 67 and 68, respectively. The RT-PCR results of 13 of these patients (9.63%) were negative, and the diagnosis of COVID-19 was made by CT scans. In 50 patients (37%), although the PCR results were positive, there was no lung finding. The non-COVID-19 group consisted of 200 individuals (age mean ± SD = 40.12 ± 10,25). One hundred of them were female (age mean = 41.51), and 100 of them were male (age mean = 38.75). To evaluate the scattergram images obtained from all individuals with and without COVID-19, patients without chronic diseases were included in the study. The demographic properties and laboratory values of the data set are given in Table 5 .
Table 5.
Demographic properties and laboratory values of the dataset.
| COVID-19, n = 135 |
Non-COVID-19 n = 200 |
|||||
|---|---|---|---|---|---|---|
| Min | Max | Interquartiles (25%–75%) | Min | Max | Interquartiles (25%–75%) | |
| AGE | 21 | 62 | 35–50 | 7 | 59 | 32–49 |
| WBC | 3.3 | 11.2 | 4.7–6.8 | 4 | 13.5 | 6.7–9.2 |
| NEU | 1.09 | 8.42 | 2.53–4.47 | 2 | 9.16 | 3.61–5.72 |
| LENF | 0.33 | 4.08 | 1.06–1.81 | 0.85 | 4.68 | 1.97–2.82 |
| MON | 0.1 | 1.96 | 0.44–0.8 | 0.064 | 1.2 | 0.47–0.71 |
| EOS | 0.001 | 0.33 | 0.01–0.09 | 0.001 | 0.87 | 0.09–0.23 |
| BAS | 0.01 | 0.1 | 0.02–0.04 | 0.001 | 0.13 | 0.03–0.06 |
| PLT | 106 | 413 | 176–244 | 143 | 435 | 220–292 |
| CRP | 1.1 | 237 | 4.02–15.2 | 1.1 | 19.7 | 1.63–5.35 |
| D-Dim | 0.1 | 3.03 | 0.25–0.58 | 0.1 | 0.93 | 0.16–0.36 |
| Ferr | 6 | 1151 | 30–139 | 7 | 378 | 18–81 |
While the blood parameters specified in Table 5 were taken from each patient, scattergram images were obtained from the WBC. First, scattergram images were classified using well-known CNN methods. In these models, the highest accuracy was determined to be 90% with the Alexnet model. In the Alexnet architecture, where the highest results were obtained, the F1 score was 87.18%, and the AUC was 89.89%. In addition, Matthews' correlation coefficient, which indicates the classification success, was 79.06%, and the sensitivity and specificity values were 89.47% and 90.32%, respectively. The percentage of healthy individuals (NPV) was 93.33%, and FPR, FDR, FNR values that should be close to zero were 9.68%, 15% and 10.53%, respectively.
A 25-depth Scat-NET architecture was developed to distinguish COVID-19 patients with higher accuracy. The results were obtained using fivefold cross validation. As can be seen from Table 4, the accuracy, Matthews' correlation coefficient and AUC values are 92.4%, 84.58% and 92.58%, respectively. The patient detection ability (sensitivity) and specificity to find healthy individuals were 92.26% and 92.9%, respectively. The percentage of healthy individuals (NPV) was 95%. The FPR, FDR, and FNR values were 7.1%, 11% and 7.74%, respectively. The CT scans of 50 of 135 COVID-19 patients were negative according to the doctor. In the Scat-NET using scattergram images, all of the patients with positive RT-PCR tests and negative CT scans were found to be positive. In addition, as shown in Fig. 7, the number of cases in which COVID-19 patients were negative was 4, 6, 4, 6 and 2. On average, 4.2 COVID-19 patients were positive. When these results are compared with the doctor's controls, the proposed method has higher accuracy than the CT scans. A literature comparison is shown in Table 6 for a better evaluation of Scat-NET and a better comparison of the results obtained.
Table 6.
Literature comparison.
| Ref. | Dataset | Model | Performance |
|---|---|---|---|
| Osman et al. (2020) | 381 scattergram images (COVID-19) | Index test and reference test | S: 85.9%, Sp:83.5 |
| Brinati et al. | WBC, 177 COVID-19. 102 Non-COVID-19 | Extremely randomized trees, DT, naive Bayes (NB), random forest (RF), SVM | S: 92%–95% Sp: 82%–86% |
| Cabitza et al. (2020) | CBC, 1624 patient, 845 COVID-19 | RF, NB, logistic regression (LR), SVM, and k-NN | AUC: 0.75–0.78, Sp: 92%–96%. |
| Ardakani et al. (2020) | 194 images CT (108 COVID-19 and 86 Non-COVID-19 l) | Resnet101, Xception | AUC:0.994, A:99.51% |
| Khan et al. (2020) | 2800 CT images, 1500 COVID-19, 1300 normal | CNN and extreme learning machine | A: 95.1%, S: 95.1%, Sp: 95%, P: 94% |
| Amyar et al. (2021) | 1369 CT images (449 COVID-19, 425 normal, 98 lung cancer, 397 other patients | Multi-task deep learning | A:94.67% |
| Aslan et al. (2020) | 219 COVID-19, 1345 pneumonia, 1341 normal | BiLSTM | A: 98.70% |
| Wu et al. (2020) | 300 CT images (150 COVID-19 and 150 Non-COVID-19 l) | Weakly supervised deep learning | S:0.833, Sp:0.956, A:90.06%, AUC: 0.943 |
| Pathak et al. (2020) | 852 CT images (413 COVID-19 and 439 non-COVID-19) | ResNet-50 and New CNN | S: 0.9146, Sp: 0.9478, A: 93.02% |
| Zhang et al. (2021) | 640 CT images (320 COVID-19 and 320 non-COVID-19) | CNN | S:93.28%, Sp:94% A:93.64% |
| Song et al. (2020) | 1485 CT images (777 COVID-19 and 708 non-COVID-19) | DRE-Net | A:86% |
| Zheng et al. (2020) | 542 CT images (313 COVID-19 and 229 non-COVID-19) | UNetþ3D deep network | A:90.8% |
| Scat-NET | 335 scattergram images (135 COVID-19, 200 Non-COVID-19 | CNN | A:92.4%, F1: %90.55 M:84.58%, AUC: 92.58% S:92.26%, Sp:92.9% P:89%, NPV:95%, FPR: 7.1% FDR: 11%, FNR: 7.74% |
Table shows similar trends to those shown by Brinati et al. (2020) and Cabitza et al. (2020), in that changes in the CBC and WBC blood parameters could be a symptom of COVID-19. Using machine learning-based algorithms, COVID-19 patients were determined to have 92%–95% sensitivity and 92%–96% specificity. In Osman's study on the diagnosis of COVID-19 from scattergram images, COVID-19 was diagnosed with statistical tests [8]. The use of CT images for suspected patients with negative RT-PCR testing is now the standard. Therefore, many researchers have developed decision support systems that assist doctors' decisions. In addition to the known CNN architectures, special CNN architectures have been designed for the diagnosis of COVID-19 disease. With these architectures, COVID-19 disease was diagnosed with over 90% accuracy from CT images. As can be seen in Table 6, there are studies where high accuracy levels of 98%–99% were obtained [24,25] as well as studies where accuracy at levels of 90–95% were obtained [19,22,23,[26], [27], [28], [29]]. The results obtained with the 25-depth Scat-NET architecture proposed in this article were successful. Parameters such as the accuracy, AUC, F1 score, sensitivity and specificity are in parallel with the literature. It can be said that a CNN model that works with a small number of images and gives good results can often give better results when it works with large data. Therefore, operations such as data augmentation are not preferred in this paper.
The results of this study showed that COVID-19 can be successfully diagnosed with the proposed Scat-NET architecture. The results show that scattergram images can be used in the diagnosis of COVID-19, and the proposed architecture can be a helpful model for expert radiologists in the diagnosis of COVID-19. We anticipate that more scattergram images will be used and that different CNN algorithms can be developed to improve the results.
One of the limitations of our study is the relatively small number of data compared to the literature. In addition, we cannot check how to treat our models in case of a large class number. In this case, overfitting may be a problem.
7. Conclusions
In this paper, CBC-DIFF parameters and scattergram images of patients admitted to the hospital with suspicion of COVID-19 were obtained. A 25-depth Scat-NET architecture is proposed to show that COVID-19 can be detected from scattergram images. This study is the first to use scattergrams and CNNs in the detection of COVID-19. The results showed that scattergram images could be used instead of costly CT scans of patients with negative suspected RT-PCR results. With our CNN model, which uses scattergram images, a different approach to COVID-19 diagnosis has been offered, and an early diagnosis and less costly solution have been developed. The model is a useful tool as a clinical decision support system to ease the burden on healthcare professionals.
Ethical approval
Ethics approval for the study protocol was obtained from the local area heath ethics committee. It is approved by the Ministry of Health of the Republic of Turkey with the form number 2021-01-13T13_56_35.
Declaration of competing interest
All authors read and approved the final manuscript. None of the authors had a conflict of interest.
References
- 1.Seghezzi M., Manenti B., Previtali G., et al. A specific abnormal scattergram of peripheral blood leukocytes that may suggest hairy cell leukemia. Clin. Chem. Lab. Med. 2018;25(5):56. doi: 10.1515/cclm-2017-0763. [DOI] [PubMed] [Google Scholar]
- 2.Gupta M., Chauhan K., Singhvi T., et al. Useful information provided by graphic displays of automated cell counter in hematological malignancies. J. Clin. Lab. Anal. 2018;32 doi: 10.1002/jcla.22392. e22392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jain R., Khurana U., Bhan B.D., et al. Mucopolysaccharidosis: a case report highlighting hematological aspects of the disease. J Lab Physicians. 2019;11(1):97–99. doi: 10.4103/JLP.JLP_126_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moradi E.V., Teimouri A., Rezaee R., et al. Increased age, neutrophil-to-lymphocyte ratio (NLR) and white blood cells count are associated with higher COVID-19 mortality. Am. J. Emerg. Med. 2021;40:11–14. doi: 10.1016/j.ajem.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selim S. Leukocyte count in COVID-19: an important consideration. The Egyptian Journal of Bronchology. 2020;14:43. [Google Scholar]
- 6.Anurag A., Jha P.K., Kumar A. Differential white blood cell count in the COVID-19: a cross-sectional study of 148 patients. Diabetes Metab Syndr. 2020;14:2099–2102. doi: 10.1016/j.dsx.2020.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou K., Li R., Wu X., et al. “Clinical features in 52 patients with COVID‐19 who have increased leukocyte count: a retrospective analysis”. Eur. J. Clin. Microbiol. Infect. Dis. 2020 doi: 10.1007/s10096-020-03976-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osman J., et al. Rapid screening of COVID-19 patients using white blood cell scattergrams, a study on 381 patients. Br. J. Haematol. 2020;2–6 doi: 10.1111/bjh.16943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitra A., Dwyre D.M., Schivo M., et al. “Leukoerythroblastic reaction in a patient with COVID‐19 infection”. Am. J. Hematol. 2020 doi: 10.1002/ajh.25793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Y., Nie H.X., Hu K., et al. Abnormal immunity of non-survivors with COVID-19: predictors for mortality. Infectious Diseases of Poverty. 2020;9(1):1. doi: 10.1186/s40249-020-00723-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Y., Zhou J., Ye K. 2020. White Blood Cells and Severe COVID-19: a Mendelian Randomization Study. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Y., Nie H.X., Hu K., et al. Abnormal immunity of non-survivors with COVID-19: predictors for mortality. Infectious Diseases of Poverty. 2020;9(1):1. doi: 10.1186/s40249-020-00723-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martens R.J.H., Van Adrichem A.J., Mattheij N.J.A., et al. Hemocytometric characteristics of COVID-19 patients with and without cytokine storm syndrome on the sysmex XN-10 hematology analyzer. Clin. Chem. Lab. Med. 2020 doi: 10.1515/cclm-2020-1529. [DOI] [PubMed] [Google Scholar]
- 14.Long C., Xu H., et al. Diagnosis of the Coronavirus disease (COVID-19): rRT-PCR or CT? Eur. J. Radiol. 2020;126 doi: 10.1016/j.ejrad.2020.108961. 108961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trisnawati I., Khair R.E., Puspitarani D.A. Prolonged nucleic acid conversion and false-negative RT-PCR results in patients with COVID-19: a case series. Annal Medic Surg. 2020;59 doi: 10.1016/j.amsu.2020.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brinati D., Campagner A., Ferrari D., et al. medRxiv; 2020. Detection of Covid-19 Infection from Routine Blood Exams with Machine Learning: A Feasibility Study”. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cabitza F., Campagner A., Ferrari D., et al. Development, evaluation, and validation of machine learning models for COVID-19 detection based on routine blood tests. Clin. Chem. Lab. Med. (CCLM) 2020:1–11. doi: 10.1515/cclm-2020-1294. [DOI] [PubMed] [Google Scholar]
- 18.Alakus T.B., Turkoglu I. vol. 140. Solitons & Fractals; 2020. “Comparison of Deep Learning Approaches to Predict Covid-19 Infection”, Chaos. 110120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pathak Y., Shukla P.K., Tiwari A., et al. IRBM; 2020. Deep Transfer Learning Based Classification Model for Covid-19 Disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Javor D., Kaplan H., Kaplan A., et al. 2020. Deep Learning Analysis Provides Accurate COVID-19 Diagnosis on Chest Computed Tomography. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou T., Lu H., Yang Z., et al. The ensemble deep learning model for novel COVID-19 on CT images. Appl. Soft Comput. 2021;98 doi: 10.1016/j.asoc.2020.106885. 106885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amyar A., Modzelewski R., Li H., et al. Multi-task deep learning based CT imaging analysis for COVID-19 pneumonia: classification and segmentation. Comput. Biol. Med. 2020;126 doi: 10.1016/j.compbiomed.2020.104037. 104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu X., Chen C., Zhong M., et al. “COVID-AL: the diagnosis of COVID-19 with deep active learning”. Med. Image Anal. 2021;68 doi: 10.1016/j.media.2020.101913. 101913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aslan M.F., Unlersen M.F., Sabanci K., et al. Applied Soft Computing; 2020. Cnn-based Transfer Learning-Bilstm Network: A Novel Approach for Covid-19 Infection Detection. 106912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ardakani A.A., Kanafi A.R., Acharya U.R., et al. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: results of 10 convolutional neural networks. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103795. 103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khan M.A., Kadry S., Zhang Y.-D., et al. Prediction of COVID-19-pneumonia based on selected deep features and one class kernel extreme learning machine. Comput. Electr. Eng. 2021;90 doi: 10.1016/j.compeleceng.2020.106960. 106960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song Y., Zheng S., Li L., et al. 2020. “Deep Learning Enables Accurate Diagnosis of Novel Coronavirus (COVID-19) with CT Images” medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S., Kang B., Ma J., et al. 2020. A Deep Learning Algorithm Using CT Images to Screen for Corona Virus Disease (COVID-19)”, medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng C., Deng X., Fu Q., et al. medRxiv; 2020. Deep Learning-Based Detection for COVID-19 from Chest CT Using Weak Label. [Google Scholar]