Abstract
The front-line imaging modalities computed tomography (CT) and X-ray play important roles for triaging COVID patients. Thoracic CT has been accepted to have higher sensitivity than a chest X-ray for COVID diagnosis. Considering the limited access to resources (both hardware and trained personnel) and issues related to decontamination, CT may not be ideal for triaging suspected subjects. Artificial intelligence (AI) assisted X-ray based application for triaging and monitoring require experienced radiologists to identify COVID patients in a timely manner with the additional ability to delineate and quantify the disease region is seen as a promising solution for widespread clinical use. Our proposed solution differs from existing solutions presented by industry and academic communities. We demonstrate a functional AI model to triage by classifying and segmenting a single chest X-ray image, while the AI model is trained using both X-ray and CT data. We report on how such a multi-modal training process improves the solution compared to single modality (X-ray only) training. The multi-modal solution increases the AUC (area under the receiver operating characteristic curve) from 0.89 to 0.93 for a binary classification between COVID-19 and non-COVID-19 cases. It also positively impacts the Dice coefficient (0.59 to 0.62) for localizing the COVID-19 pathology. To compare the performance of experienced readers to the AI model, a reader study is also conducted. The AI model showed good consistency with respect to radiologists. The DICE score between two radiologists on the COVID group was 0.53 while the AI had a DICE value of 0.52 and 0.55 when compared to the segmentation done by the two radiologists separately. From a classification perspective, the AUCs of two readers was 0.87 and 0.81 while the AUC of the AI is 0.93 based on the reader study dataset. We also conducted a generalization study by comparing our method to the-state-art methods on independent datasets. The results show better performance from the proposed method. Leveraging multi-modal information for the development benefits the single-modal inferencing.
Keywords: COVID-19, Multi-modal, Artificial intelligence, Reader study
1. Introduction
Coronavirus disease 2019 (COVID-19) is extremely contagious and has become a pandemic [1], [2]. It has spread inter-continentally in the third and forth wave [3] and suspected to be currently entering the second wave [4], [5], [6] in various countries, having infected more than 30 million people and caused nearly 3.5 M deaths till May 2021 [7]. The mortality rate of this disease differs from country to country ranging from 2.5% to 7% compared with 1% from influenza [8], [9], [10]. Considering different age groups, elder people and patients with comorbidities are most vulnerable and more likely to progress to a life-threatening condition[11]. To prevent the spread of the disease, different governments have implemented strict containment measures [12], [13] which have aimed to minimize transmission. Because of the strong infection rate of COVID-19, rapid and accurate diagnostic methods are urgently required to identify, isolate and treat the patients specially considering that effective vaccines are still under development.
The diagnosis of COVID-19 relies on reverse-transcriptase-polymerase chain reaction (RTPCR) test [14], [15], [16]. However it has several drawbacks. The RTPCR tests often require 5 to 6 h to yield results. Sensitivity of RTPCR depends on the stage of the infection [16] and can be as low as 71%[17]. Therefore care must be taken on interpreting RTPCR tests. More importantly, the cost of RTPCR prevents large population from being tested in the developing and highly populated economies.
From the imaging domain, chest CT may be considered as a primary tool for COVID-19 detection [18], [15], [19] and the sensitivity of chest CT can be greater than that of RTPCR (98% vs 71%) [17], but the cost, time and risks of imaging including dose and need for system decontamination can be prohibitive in most markets. In contrast, portable X-ray (XR) units are accessible, cost and time effective, with lower radiation dose and thus the defacto imaging modality used in diagnosis and disease management. Since X-ray imaging has limited capability to provide detailed 3D structure of anatomy or pathology of chest cavity, it is not regarded as an optimum tool for quantitative analysis [20]. Also due to the imaging apparatus and nature of the X-ray projection, it is challenging for radiologists to identify relevant disease regions for accurate interpretation and quantification[21], [22]. The early detection of COVID-19 through XR images is particularly challenging, even for expert radiologists, as it cause only subtle changes in the projected image.
To alleviate the lack of experienced radiologists and minimize human effort in managing an exponentially growing pandemic and the impending task to triage suspected COVID-19 subjects, the academic and industry communities have proposed various systems for diagnosing COVID-19 patients using X-ray imaging [23], [24], [25], [26]. The performance of some AI systems to detect COVID-19 pneumonia was comparable to radiologists to identify presence or absence of COVID-19 infection[24]. The common outputs of these diagnostic solutions or triage systems are classification probabilities for COVID-19 and non-COVID-19 accompanied by heatmaps localizing the suspected pathological region or areas of attention.
Artificial intelligence algorithms can be used to discern subtle changes and aid radiologist analysis in various applications.[27], [28], [29], [30], [31], [32], [33], [34], [35] Although the performance of a host of systems approaches the level of radiologists for chest X-rays classification, very limited studies have verified the detection and segmentation of the disease regions compared to human annotation on X-rays. Segmentation is critical for (a) severity assessment of the disease; and for (b) follow-up for treatment monitoring or progression of patient condition. Although human annotations can be obtained for COVID-19 regions on X-rays, the certainty of regions is weak compared to annotations derived from CT. The development in the field suffers from the lack of availability of COVID-19 X-ray images with corresponding region annotations. The contribution of our work builds on establishing a multi-modal protocol for our analysis and downstream classification of X-ray images.
The core of this study is the transformation between multi-modal data. Previously, [36] has shown that convolutional neural networks trained on propagated MR contours significantly outperform those trained on CT contours and also experts contouring manually on CT for hippocampus segmentation. This is probably due to the poor visibility of hippocampus on CT. For COVID-19 application, [37] leverages existing annotated CT images to generate frontal projection X-ray images for training COVID-19 chest X-ray models. Their model far outperforms baselines with limited supervised training and may assist in automated COVID-19 severity quantification on chest X-rays. Barbosa Jr. et. al. [38] have also leveraged CT ground-truth for generating synthetic X-ray where their approach takes ratios of disease region to lung region using both synthetic X-ray (SXR) and regular X-ray images and force them to be equal to the ratio of disease volume to lung volume in CT. It might be challenging to incorporate such a definition into clinical practice. Our deep learning model also trains using data from CT, by generating synthetic X-ray from paired CT scans to complement the including original X-ray images. At the inference end of the pipeline, only X-ray images are exclusively used. Differently from existing studies, we established a synthetic X-ray generation scheme to generate a multitude of realistic synthetic X-ray to significantly augment X-ray images to expand our training pool. Second, we use synthetic X-ray as a bridge to transfer the ground-truth from CT to the original X-ray geometry.
Another contribution of our study is to highlight the serious domain shift issue when collecting images from multiple data sources where all three-class image data from the same source is not always available for training and testing.
Although a preliminary work of this study has been previously published as a conference paper[39], this paper is substantially different from preliminary work in that it presents a comprehensive study of proposed approach and includes an observer study validation.
2. Method
2.1. Solution Design Overview
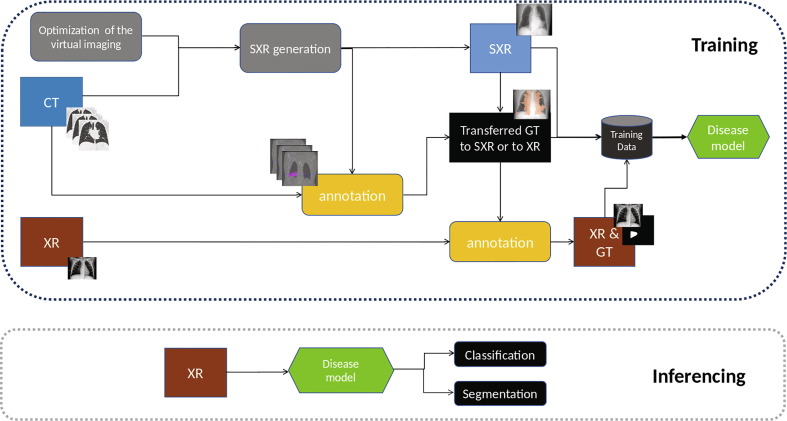
The key idea is to learn the disease patterns jointly using multi-modal (CT and XR) data but to inference the solution using only a single modality (XR). In this study, we are not particularly focusing on the design of deep-learning networks, but rather on improving data sufficiency and enhancing ground-truth quality to improve the visibility of abnormal tissue on a lower dimensional modality. In order to maximally leverage available X-ray data, CT scans and paired X-ray and CT images, we have designed a pipeline as illustrated in Fig. 1 . Disease regions in X-ray and CT are annotated independently by trained staff with varying levels of experience. Using CT, we generate multiple SXR image per CT volume and the corresponding projected 2D masks of the diseased region(Section 2.2). For patients where X-rays and CTs (paired XR and CT data) are acquired within a small time-window (48 h given that COVID-19 is a fast evolving disease and pathology regions in the lungs may change dramatically between exams), we automatically project and transfer the CT masks using the SXR to match the corresponding XR. The transferred annotations on XR are further adjusted manually reviewed by trained staff to build our pristine mask ground-truth/annotations (Section 2.3). For patients where the time-window between CT and XR exams is larger than 48 h, no ground-truth transfer is performed across the two modalities, but data is still added in the training pool. With all available XR and SXR images along with corresponding disease masks, we train a deep-learning convolutional neural network for diagnosing and segmenting COVID-19 disease regions on X-ray.
Fig. 1.
The training scheme and inferencing design.
2.2. Synthetic X-ray Generation
Original X-ray images are generated by shining an X-ray beam with initial intensity on the subject and measuring the intensity (I) of the beam having passed through an attenuating medium at different positions using a 2D X-ray detector array. The attenuation of X-ray beams in matter follows the BeerLambert law, stating that the decrease in the beam’s intensity is proportional to the intensity itself (I) and the linear attenuation coefficient value() of the material being traversed. (1).
| (1) |
From this we can derive the beam intensity function over distance to be an exponential decay as the following:
| (2) |
In medical X-ray images the values saved to file are proportional to the expression seen in 3, which is actually a summation of the attenuation values along the beam. If the subject is made up of smaller homogeneous cells, this could be formulated as a sum.
| (3) |
3D CT images are constructed from many 2D X-ray images of the subject taken at different angles and positions (helical or other movement of the source-detector pair) by using a tomographic reconstruction algorithms such as iterative reconstruction or the inverse radon transform. The voxel values of the constructed CT image are the linear attenuation coefficient values specific to the used X-ray beam’s energy spectrum. These values are converted to Hounsfield unit (HU) values as seen in Eqn. 4, where and are the linear attenuation coefficient for water and air respectively for the given X-ray source. Using this unit of measure transforms images taken with different energy X-ray beams to have similar intensity values for the same tissues, scaling water to 0 HU, air to −1000 HU.
| (4) |
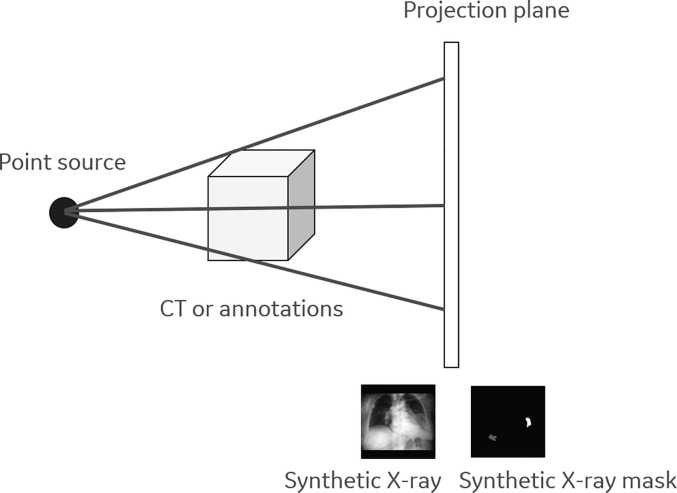
An inverse process can help create X-ray images from CT images by projecting the constructed 3D volume back into a 2D plane along virtual rays originating from a virtual source. Since the X-ray image (Eqn. 3) is an integral/summation of attenuation values along the projection rays, established methods create projection images by converting the CT voxel values from HU to linear attenuation coefficient and simply summing the pixel values along the virtual rays (weighted by the travelled path-length by the ray withing each pixel). To achieve realistic pseudo X-ray we used a point source for the projection which accurately models the source of the X-ray, as in actual X-ray imaging apparatus. For the projection itself we used the ASTRA-toolbox[40], which allowed parameterization of the beam’s angle (vertical and horizontal) as well as the distances between the virtual point source, the origin of the subject and the virtual detector (see Fig. 2 ). As X-ray images have the same pixel spacing in both dimensions, while CT images usually have larger pixel spacing in the z direction, we re-sampled the 3D volumes to have isotropic (0.4 mm) spacing prior to computing the projection. In clinical X-ray images, the radiation source is behind and the detector is in front of the patient - also known as posterior-anterior (PA) view the images generated appear to be flipped horizontally. To conform with this protocol, we also flipped PA projections. For each CT volume we perform projections at different angles. The range of angles between the center of the CT image and the virtual X-ray point source falls between −10 and 10 degrees around the longitudinal axis, and −5 and 5 degrees around the mediolateral axis of the patient. Based on what is traditionally used in clinical practice, we used 2 distance settings between the center of the CT volume and the virtual X-ray source, 1 m and 1.8 m, while the distance between the CT volume center and the virtual detector is set to 0.25 m.
Fig. 2.
The illustration of synthetic X-ray and its mask generation.
It is important to point out that in case of small field-of-view CT images, where the patient doesn’t fit into the reconstruction circle in the horizontal plane, creating coronal or sagittal projection will not result in lifelike X-rays image as body parts are missing from the virtual ray’s path.
During our work we also projected the segmentation ground truth masks for CT images to 2D format. The same transformations were applied to these binary masks as to the corresponding CT volumes. In our workflow we use both a binary format of the 2D projected masks - created by setting all non-zero values to 1 - and a depth-mask format which retains information of the depth of the original 3D mask. The latter was scaled so the values have the physical meaning of depth.
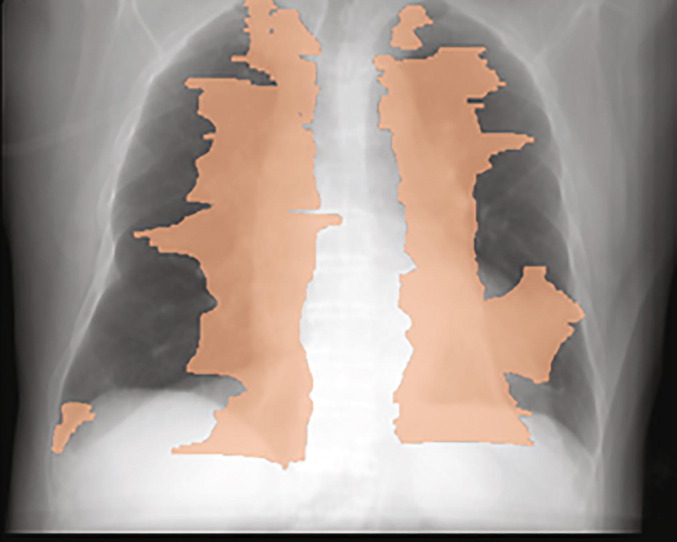
It should be noted that one important novelty in our study is that our segmentation/groundtruth is based on CT groundtruth which means that the disease region are from 3D. If we project the 3D disease on X-ray, the disease can be outside of traditional defined 2D X-ray lung while the real lung is much bigger than the black region on X-ray (see Fig. 3 ).
Fig. 3.
A synthetic X-ray with its corresponding projected disease mask as an overlay.
2.3. Pristine Annotation Generation
For paired XR and CT images of the same patients, our objective is to transfer the pixel-wise ground-truth from CT to SXR and from SXR to XR. SXR serves as a bridge between CT and XR.
For each CT volume, we generate a number of SXRs and corresponding disease masks by varying the imaging parameters as mentioned in Section 2.2. The goal is to register each SXR candidate to XR so that the disease region between SXR and XR have the best alignment so that we can apply the same transformation on the disease mask from SXR and generated registered disease mask for the paired XR. As for the same CT, multiple SXRs are generated resulting different registered masks and we simply select the one with the best mutual information between registered SXR and XR. Another approach to select optimal SXR involves comparison of the lung mask region. The pair with the most overlapping mask was used to select the ideal pair.
To register SXR to XR, one issue to address is that fields of body view are different between the two and SXR. Furthermore SXR images manifest with hands and arms up while the XR images have the hands and arms hanging downward, normal position of the body. To alleviate this problem during image registration, instead of using original images, we generate a lung region of interest (ROI) image by applying lung segmentation on synthetic X-ray and X-ray. The lung segmentation solution is an inhouse developed method using a 2D U-Net [41] trained previously using a different dataset outside the scope of this study. We use affine registration to maximize the mutual information between lung ROIs from SXR and XR. Once registration error converges or registration step limit is reached, the obtained transformation is applied to the annotations corresponding to the SXR to generate transferred annotations.
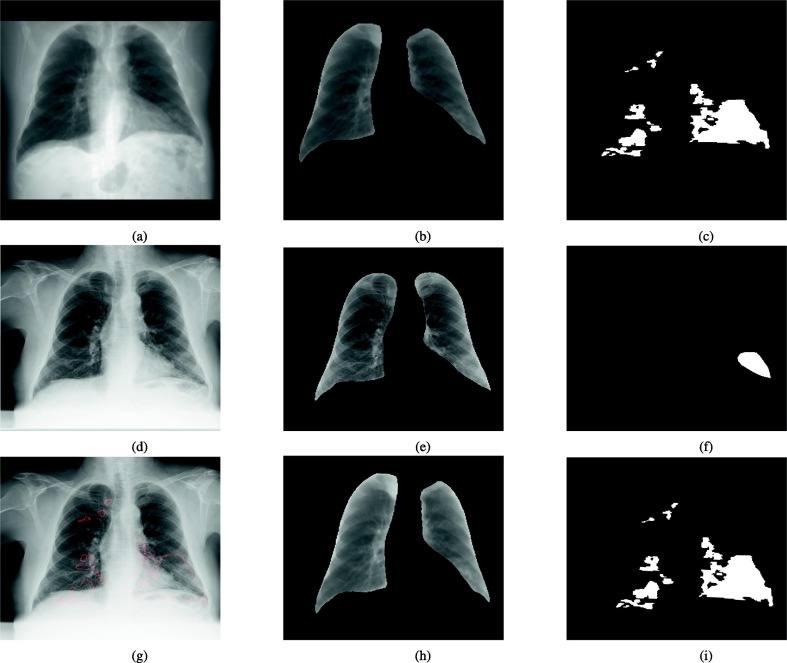
Fig. 4 shows one example how we transfer the CT ground-truth from CT to synthetic X-ray and from synthetic X-ray to real X-ray. We observe that there are moderate differences between annotations from original X-ray and those annotations derived from transformed and transferred CT data implying that the visibility of COVID-19 related pneumonia on X-ray may not be ideal and most comprehensive.
Fig. 4.
An example of transferring synthetic X-ray mask to X-ray mask. Top: a representative synthetic X-ray generated from CT, the corresponding lung image and disease mask; Middle: paired X-ray, the corresponding lung image and direct disease annotation from X-ray; bottom: X-ray with transferred annotations from CT shown as red contour; registered lung image from synthetic X-ray and transferred disease annotations from synthetic X-ray.
Although we have an approximate spatial match between SXR and XR, we cannot blindly use the transferred ground-truth(GT) for XR for training. The disease could have rapidly evolved during the first and second scan even within 48 h. In our study, the transferred groundtruth can only be used as a directional guidance by the human expert to help the annotation of the 2D X-rays. The general instruction to the annotators is to disallow/erase any transferred regions when no underlying lesions are visible on the X-rays image, and keep minimally visible regions, even if they appear in the heart/diaphragm region.
2.4. COVID-19 Modeling and Evaluation Strategy
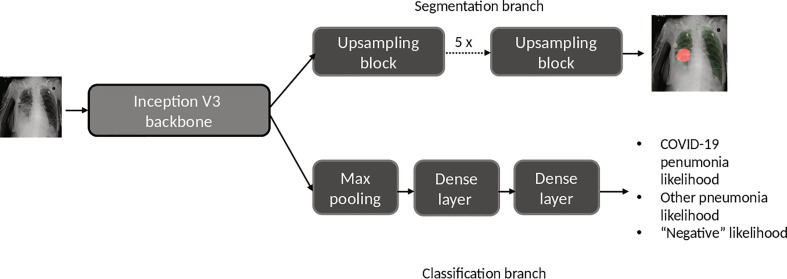
During this extra-ordinary COVID-19 pandemic, AI systems have been investigated to identify anomalies in the lungs and assist in the detection, triage, quantification and stratification (e.g. mild, moderate and severe) of COVID-19 stages. To help radiologists do the triage, our deep-learning model takes frontal (anterial-posterior view or posterior-antierial view) X-ray as input and outputs two types of information (see Fig. 5 ): (i) location of the disease regions and (ii) classification. For location, the network generates a low resolution (480 x 480) segmentation mask to identify disease pixels on XR and SXR images related to both COVID-19 and regular pneumonia cases. For classification, a fully connected neural network (FCN) outputs the probablity of (i) COVID-19, (ii) regular pneumonia or (iii) a negative finding for each given input AP/PA XR image.The classification branch consists of one maxpooling layer, a dense layer of 10 nodes and a dense layer of 3 nodes. The segmentation branch consists of 5 upscale blocks and each block consists of one residual block and one transpose convolution layer. Each upscale block doubles the dimension of each of the feature channels. Both XR and SXR images are first resized to 1024 x 1024 pixels and normalized by Z-score method before being fed into the model. In this study, we form COVID-19 disease classification as a three-class classification problem hypothesizing that the distribution of abnormality in the lung may become a differentiator between COVID-19 and regular pneumonia patients. But it should also be noted that a three-class classification can be converted to a two-class classification when inferencing by taking the maximum probability between the COVID-19 disease and pneumonia classes. For training, we use a combination of cross-entropy loss from the classification branch and Dice loss from the segmentation branch. The model is trained for 50 epochs, with batch size 3 and using the Adam optimizer[42].
Fig. 5.
The schematic overview of our proposed classification and segmentation deep-learning model.
Many publications have released their algorithms in open online forums or are marketing the same as additional pneumonia indicators. However, the robustness of the algorithms and their clinical value is somewhat unproven. A few studies have characterized systems for COVID-19 prediction with stand-alone performance that approaches that of human experts. However, all the existing works have either no established pixelwise ground-truth or are evaluated using pixel-wise ground-truth from purely X-ray annotations with uncertainties from annotators.
Our deep learning model was trained with and without the multi-modal data from CT cases to investigated the benefits of multi-modal learning. We evaluated our approach in three separate aspects. First, AI model predictions were compared for the accuracy of the image-level classification labels (COVID-19 pneumonia, other pneumonia or negative). Second, the model segmentation of disease regions for the COVID-19 class was evaluated against direct human X-ray pixel-wise pathology annotations/masks. Third, the model segmentation of disease regions for the COVID-19 pneumonia was compared against human pristine pixel-wise annotations/masks.
3. Data and Groundtruth
In this study, we formed a large experimental dataset consisting of real X-ray images and synthetic X-ray images originating from CT volumes. The data was sourced from in–house/internal collections as well as publicly available data sources including Kaggle Pneumonia RSNA [43], Kaggle Chest Dataset [44], PadChest Dataset [45], IEEE github dataset[46], NIH dataset [47]. Any image databases with limiting non-commercial use licenses were excluded from our train/test cohorts. Representative paired and unpaired CT and XR datasets from US, Africa, and European population were included and were sourced through our data partnerships.
Outcomes were derived from information aggregated from radiological and laboratory reports. A summary of the database and selected categories used for our experiments is summarized in Table 2 where the in–house test dataset is a subset of the complete test dataset. As the trained model will be inferenced on real X-ray images, we remove synthetic X-rays from our validation and testing cohorts. A dedicated in–house testing dataset has been used for our study due to the availability of complete ground truth on these cases marked into three classes: COVID-19, regular pneumonia and negative. From the general testing datset, some of the data sources do not contain all three-class images. This may cause domain shift-based bias. Table 1 shows the details image composition of different data sources.
Table 2.
Dataset breakdown for our experiments.
| Dataset | X-rays (# XMA, # PMA) | Synthetic X-rays (# SMA) |
|---|---|---|
| train COVID-19 | 974 (247, 77) | 21487 (8322) |
| train pneumonia | 10175(6108, 17) | 11312 (5380) |
| train negative | 14859 (NA,NA) | 12542 (NA) |
| val COVID-19 | 113 (37,2) | NA |
| val pneumonia | 531 (473,8) | NA |
| val negative | 3301 (NA, NA) | NA |
| test COVID-19 | 307 (68, 52) | NA |
| test pneumonia | 1006 (345,33) | NA |
| test negative | 2271 (NA,NA) | NA |
| in–house test COVID-19 | 266 (68,52) | NA |
| in–house test pneumonia | 116 (45,33) | NA |
| in–house test negative | 37 (NA,NA) | NA |
Table 1.
Data source details.
| Data source | COVID-19 (train/val/test) | Pneumonia (train/val/test) | Negative (train/val/test) |
|---|---|---|---|
| Kaggle Pneumonia RSNA | NA | 5412/300/300 | NA |
| Kaggle Pneumonia Chest | NA | 3875/8/390 | 1341/8/234 |
| PadChest Dataset | NA | 694/200/200 | 4925/2000/2000 |
| IEEE github dataset RSNA | 122/29/41 | NA | NA |
| NIH dataset | NA | NA | 6018/757/0 |
| In–house negative data source | NA | NA | 2379/497/0 |
| In–house three class source | 852/84/266 | 194/23/116 | 196/39/37 |
Two levels of groundtruth are associated with each image: image-level groundtruth and pixel-level (segmentation) groundtruth.
For image-level groundtruth, each image is assigned with a label of COVID-19, pneumonia or negative. All in–house X-rays and CTs and the COVID-19 images from the public data sources were confirmed by RTPCR tests. The labels of pneumonia and normal images from public data sources are given by radiologists.
For the pixel-wise/segmentation groundtruth (masks), we have four types of annotations: (i) X-ray manual Mask Annotations (XMA) made by annotators purely based on X-rays without any information from CTs; (ii) Synthetic Mask annotations (SMA) generated by the projection algorithms based on CT annotations for synthetic X-rays; (iii) Transferred CT Mask Annotations (TMA) automatically generated by registration algorithms which transfer annotations from CT to X-ray using SXR as a bridge; (iv) Pristine Mask Annotations (PMA) generated by trained human annotators with the adjustment to the TMA. The voxel/pixel-wise annotations from CT and X-ray except for RSNA dataset were performed by internal annotators. The RSNA pixel annotations were generated by fitting ellipses to the bounding boxes provided from the data source.
4. Experiment Settings
To show the benefits of multi-modal training for developing COVID-19 model, we have conducted training with 4 different training datasets summarized in Table 3 where S3 contains paired images used twice with XMA and PMA.
Table 3.
Different training sets.
| training dataset setting | Description |
|---|---|
| S1 | X-ray images with XMA |
| S2 | S1 + synthetic X-ray images with SMA |
| S3 | S1 + X-ray images with PMA |
| S4 | S1 + synthetic X-ray images with SMA + X-ray images with PMA |
To further improve the robustness of learning using a single model, we also conduct ensemble learning of three classifiers with different weight initlizations of the same training data setting (S4). The output of the ensemble is the averge of the outputs from three trained models.
5. Reader Study
To assess the usability of this AI system and justify its performance, we have conducted a reader study. Two certified radiologists are invited to read 50 X-ray images (26 COVID-19, 8 pneumonia and 23 negative). Both radiologists have over 10 years’ experience on thoracic imaging.Radiologists performed pixelwise annotations for the disease regions in addition to classifying them. For the classification, five options with increasing suspiciousness levels were chosen: no pathology at all, no pneumonia sign, nonCOVID-19, indeterminate COVID-19 and probable COVID-19.
5.1. Evaluation Metrics
To evaluate the performance of our inferred classification on the test subjects, we used the area under the receiver operating characteristic curve (AUC) between different combinations of positive and negative classes including COVID-19 pneumonia vs other pneumonia, COVID-19 pneumonia vs other pneumonia + negative and COVID-19 pneumonia vs negative for different deployment scenarios.
Dice coefficient was used to evaluate the exactness of our pathology localization,
6. Results
6.1. Pristine annotation creation
With three different pixel-wise annotations on X-ray, we evaluated overlap between XMA, PMA and TMA.
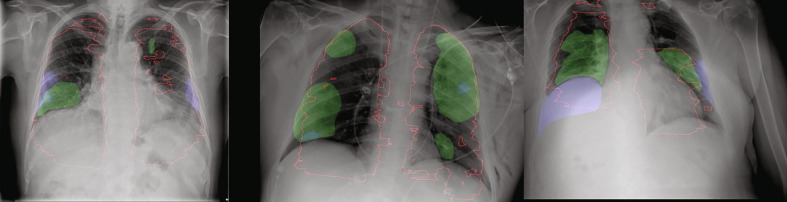
We can observe (see Table 4 ) that after using TMA (transferred CT annotations), the consistency of human annotations to CT annotations is largely improved from 0.28 to 0.47 in terms of Dice coefficient. The Dice coefficient between XMA (X-ray manual mask annotations) and PMA (pristine mask annotations) are also moderate which means PMA has both good consistency to TMA and XMA while the consistency between TMA and XMA is poor. Fig. 6 shows a number of examples with TMA, XMA and PMA. The X-ray annotations show large inconsistency with automated CT transferred annotations.
Table 4.
Area overlapping between different annotations.
| Comparisons | Dice |
|---|---|
| XMA vs TMA | 0.28 |
| PMA vs TMA | 0.47 |
| XMA vs PMA | 0.50 |
Fig. 6.
Examples with large annotation inconsistencies where TMA as red contour, XMA as blue regions and PMA as green regions.
6.2. Model Evaluation
The evaluation of the model is performed in terms of both classification and segmentation of COVID-19 disease regions. To test the effect of domain shift, we show the evaluation results on both all X-ray test dataset and in–house test dataset where COVID-19 regular pneumonia and normal cases are all available.
Regarding classification, Table 5 shows AUC based on different combinations for positive and the negative classes. By adding synthetic X-rays, the AUC increases for COVID-19 pneumonia vs other pneumonia + negative increase from 0.89 to 0.93. The addition of adding pristine groundtruth does not further increase the AUC. The same increase is observed if AUC is computed using COVID-19 pneumonia as positive and other pneumonia as negative class. We formulate the triaging of COVID-19 patients as three-classification problem to cope with different application situations. When inferencing, clinicians can adjust AI outputs depending on the different use-cases. For example, if clinicians want minimum regular pneumonia and negative patients in the recall, the probablity from COVID-19 is a sufficient indicator. If clinicians prefer high sensitivity, and recalling regular pneumonia patients is not considered a clinical burden, the maximum among probablity of COVID-19 class and pneumonia class from our solution can be considered as an indicator.
Table 5.
Classification results: AUC measures of different training schemes on different datasets with different positive and negative compositions.
| Training dataset setting vs test AUC | AUC C vs P + N on XR testset |
AUC C vs P on XR testset |
AUC C vs P + N on in–house XR testset |
AUC C vs P on in–house XR testset |
|---|---|---|---|---|
| S1 | 0.98 | 0.98 | 0.89 | 0.87 |
| S2 | 0.99 | 0.98 | 0.93 | 0.92 |
| S3 | 0.99 | 0.98 | 0.91 | 0.90 |
| S4 | 0.99 | 0.99 | 0.93 | 0.92 |
| S4 ensemble | 0.99 | 0.99 | 0.93 | 0.93 |
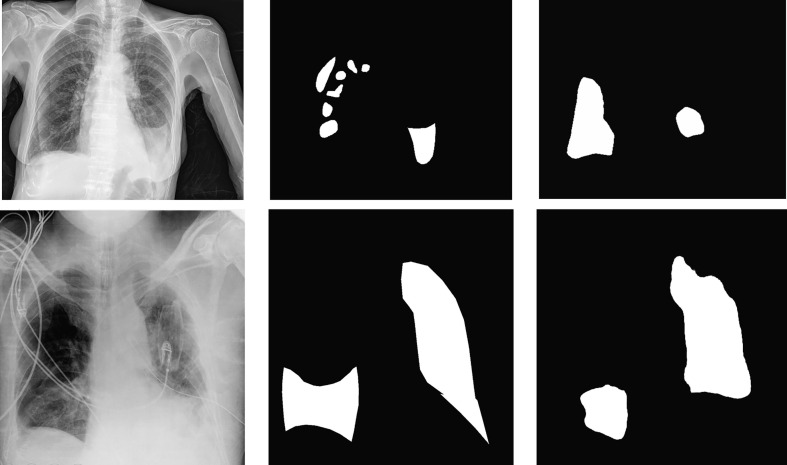
We measure the Dice coefficients to estimate the segmentation accuracy. As PMA were obtained for the in–house dataset, we have measured Dice on all testing images with XMA, in–house test images with XMA and testing images with PMA only shown in Table 6 . Adding PMA can largely improve the Dice measures across different test settings pushing it up to 0.70. Fig. 7 shows examples of AI detection and segmentation of COVID-19 regions with manual annotations as overlay as well in one uni-lateral and one bilatral case. In the bilateral case, our AI missed the consolidation in the bottom of the left lung. The AI might recognize this consolidation as pleural effusion and therefore dismissed this region.
Table 6.
Segmentation results: Dice measures of different training schemes on different datasets.
| Train dataset setting vs test Dice | XR testset with XMA | in–house XR testset with XMA | in–house XR testset with PMA |
|---|---|---|---|
| S1 | 0.58 | 0.59 | 0.59 |
| S2 | 0.57 | 0.56 | 0.58 |
| S3 | 0.60 | 0.62 | 0.70 |
| S4 | 0.57 | 0.58 | 0.62 |
| S4 ensemble | 0.58 | 0.59 | 0.64 |
Fig. 7.
Segmentation examples where images on the left are original X-ray images, in the middle are PMA and on the right are AI segmentations.
6.3. Reader Study Results
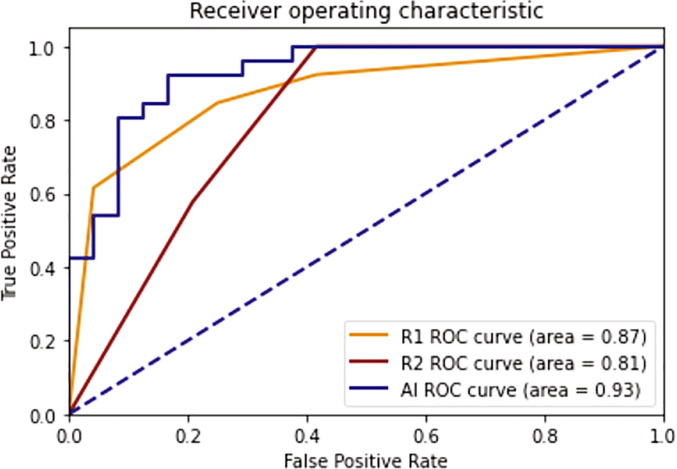
From the reader study, from the detection/segmentation perspective, the AI has shown good consistency with respect to radiologists (see Table 7 ) considering that the DICE between two radiologists on the COVID group is 0.53. From a classification perspective, the AUCs of two readers are 0.87 and 0.81 while the AUC of the AI is 0.93 for this reader study dataset. Fig. 8 shows ROC curves of radiologists and the AI system in the use case of COVID versus non-COVID classification.
Table 7.
Area overlapping between readers and AI.
| Comparisons | Dice |
|---|---|
| Radiologist 1 vs Radiologist 2 | 0.53 |
| Radiologist 1 vs AI | 0.52 |
| Radiologist 2 vs AI | 0.55 |
Fig. 8.
ROC curves of radiologists and the AI system in the use case of COVID-19 versus non-COVID-19 classification.
6.4. Generalization Study
To evaluate the generalization of the proposed method, we considered few state-of-the-art methods and puclic datasets and we presented our anlaysis in this section. Since the current models are mostly focusing on one task, i.e. segmentation or classification only, two methods are compared to the proposal in different perspective. For segmentation task, CT2X-ray method [37] is considered, which employs the CT to X-ray transformation as the multi-modality approach for model training in X-ray domain. In addition, to validate the classification performance, Covid-Net [48] is also adopted for the comparison.
To perform a fair comparison, extra public datasets are used for the above segmentation and classification tasks. For segmentation task, BIMCV dataset [49] is adopted, which provides 14 X-ray images with carefully annotated ground truth (excluding lateral views to fit our method). As for classification task, 300 positive COVID-19 X-ray images are obtained from [50] (by carefully excluding the RSNA images to avoid information leakage). Similarly, 300 negative non-COVID images are obtained from CheXpert dataset [51]. Note that these 300 + 300 images are obtained from the first 300 images of each dataset.
The segmentation results for two different multi-modality methods are evaluated using Dice score, which are 0.57 and 0.51 for the proposed method and CT2X-ray method, respectivaly. For the classification results, the proposed method is also better than Covid-Net when evaluating on the independent datasets. These results are shown in the Table 8 . There is a drop noticed on the classification performance and the cause is attributed to the extremely low resolution (256x256) of the available.
Table 8.
Classification performance on independent dataset.
| Comparisons | AUC | Specificity | Sensitivity |
|---|---|---|---|
| Covid-Net | 0.55 | 0.55 | 0.55 |
| Proposed | 0.81 | 0.73 | 0.72 |
7. Conclusion and Discussion
In this study, from multi-modal perspective, we have developed an artificial intelligence system which learns from a mix of high dimensional modality CT and X-ray but inferences only on low dimensional mono-modality X-ray for COVID-19 diagnosis and segmentation/localization of the diseased regions. The system classifies a given image into three categories: COVID-19, pneumonia and negative. We show that by learning from CT, the performance of the AI system seems to improve both classification and segmentation of the pathology. Our AI system achieves a classification AUC of 0.99 and 0.93 between COVID-19 pneumonia and other pneumonia plus negative on the full testing dataset and the subset in–house dataset, respectively. The Dice of 0.57 and 0.58 are obtained for COVID-19 disease regions on full testing dataset and the subset in–house dataset using X-ray direct annotations, respectively. The Dice is increased to 0.62 when pristine ground-truth transferred from CT is used for training and testing. We also observe that with ensemble modelling, the classification performance and segmentation performance can be further improved over a single S4 model. We have also conducted a reader study to justify the performance of AI. The DICE of AI (0.53–0.55) is at a comparable level of radiologist (0.53) and AI outperformed two radiologists on the triaging of the COVID19 patients. Accurate classification can aid physician in triaging patients and make appropriate clinical decisions. Accurate segmentation enhanced confidence in the triage, and helps in quantitative reporting of disease for reporting and monitoring progression.
The transferred groundtruth from CT is used as hint for guiding the annotation and generating pristine markings labeling the data and pathology. One can imagine TMA as computer-aided detection (CAD) markers to aid the radiologists to improve the disease region detection/delineation. In the mainstream FDA or CE reader studies[52], [53], it is often mentioned that CAD markers are used in a similar way to improve the accuracy and consistency of the disease detection. Learning from a second modality (CT) in our multi-modal approach has two main implications. One impact is to add synthetic X-ray and corresponding disease masks with different projection parameters to significantly augment training image pool and ensure data diversity. Another benefit relates to additional pathology evidence from a imaging modality with higher-sensitivity observed when we transfer the CT annotations to original X-ray using synthetic X-ray as a bridge, thereby allowing manual adjustment used for training. The first addition contributes mostly towards gain in the classification accuracy and the second addition contributes substantially to the gain in disease localization. Although the CT resolution is generally lower than X-ray resolution, the diagnosis of COVID-19 disease depends on disease distribution over the left and right lungs. We found limited benefit on pathology segmentation by adding synthetic X-rays (S2) compared to leveraging only real X-rays (S3) with pristine ground-truth. Such differences may be due to the fact that synthetic X-rays have different intensities and contrast compared to real X-rays and segmentation annotations of synthetic X-ray are derived directly from CT where some lesions may not be visible in the original X-rays. In S2 setting, the mask annotations for synthetic X-rays come directly from CT annotations without manual adjustments. The mask annotations can be inconsistent to visual perception of abnormality of synthetic X-rays. This may leads to less benefits on the segmentation from S2 setting. The quality of the synthetic X-ray may play an important role and is worth further investigation, perhaps using generative networks to make the synthetic X-ray more realistic.
Our study shows that there exists substantial inconsistency between X-ray direct annotations and automated transferred CT annotations. When using transferred annotations as hints, the second version of X-ray annotations (pristine groundtruth) are more consistent to automated transferred CT annotations. It also indicated that the automatically transferred CT annotations cannot be directly used for training as some lesions visible in CT are just not visible in X-ray and also because of the disease change between the two exams. The Dice metric between direct annotations and pristine groundtruth is 0.50 which might be a good reference indicating the entitlement of AI based segmentation. One important difference between XMA and TMA is that usually radiologists tend to annotate lung disease regions just within the dark areas which are assumed to be lung parenchyma on X-ray. However, our Fig. 3 clearly shows that after projecting the 3D disease from the CT lung images, the disease can be actually outside of dark lung region like the cardiac region. Readers can further adjust their annotations to cover possible and visible disease regions. We do not plan to use the TMA directly as some of projected regions are obscured on X-ray images. Readers can also just adjust annotations by only looking at CT annotations for guidance. However, we aim for pixel-level annotations, therefore using the transferring approach, TMA is generated for pixel-level guidance for the manual operators. SMI also supplements the training cohort along with the synthetic X-ray as an augmentation opportunity.
It should be further noted that the automated transferred annotations can not be directly used in training as registration error can cause incorrect definition of pathology.We recognize that COVID-19 is a fast changing disease and since we are pairing X-ray and CT acquired within a pre-defined time-window of 48 h, the disease status in CT might be quite different compared to the disease status when X-ray is taken. Therefore manual adjustment is an important consideration.
One important observation we would like to point out is that domain shift can occur when developing an AI solution using data collected from different data sources espeically for a fast envolving disease or when there are extraordinary limitations preventing access to large volumes of data. This also confirms the observation by DeGrave et al. [54]. It was shown that recent deep learning systems to detect COVID-19 from chest radiographs rely on confounding factors rather than medical pathology, creating an alarming situation in which the systems appear accurate, but fail when tested in new hospitals. Excellent performance is achieved in a general testing dataset but prominent performance drop is observed in the results on in–house testing set. On one hand, the COVID-19 and the pneumonia cases are confirmed with RTPCR tests, while RTPCR test has lower sensitivity making the ground-truth less dependable. On the other hand, the public datasets do not contain all three-classes of images together from the same source thereby confounding the trained model to recognize both disease and data source at the same time for the classification task. Although both normalization and extensive augmentations are applied to balance the data pools during training, when this model is tested on the general testing dataset, the recognized data-source may help to achieve an unrealistically good classification results due to the data-source bias.
In the research and industry community, efforts have been made to apply AI into imaging-based pipeline of for the COVID-19 applications. However, many existing AI studies for segmentation and diagnosis are based on small samples and based on single data source, which may lead to the over-fitting of results. To make the results clinically applicable, a large amount of data from different sources shall be collected for evaluation. Moreover, many studies only provide classification prediction without providing segmentation or heatmap which makes AI systems lack explainability. By providing the segmentation, we aim to fill this void, enhancing the promotion of AI in clinical practice. On the other hand, the imaging-based diagnosis has limitations and clinicians make the diagnosis considering clinical symptoms also. An AI system can be largely enhanced with incorporation of patient clinical parameters [55] such as blood oxygen level, body temperature, to further enhanced the capability to accurately diagnose pathological conditions.
WHO has recommended[56] a few scenarios where chest-imaging can play an important role in care delivery. From the triage perspective, WHO suggests using chest imaging for the diagnostic workup of COVID-19 when RTPCR testing is not available (timely) or is negative while patients have relevant symptoms. In this case, the classification support from our AI can aid radiologists to identify COVID-19 patients. From monitoring or temporal perspective, for patients with suspected or confirmed COVID-19, WHO suggests using chest imaging in addition to clinical and laboratory assessment to decide on hospital admission versus home discharge, to decide on regular admission versus intensive care unit (ICU) admissions, to inform the therapeutic management. In these scenarios, accurate segmentation of disease regions is essential for the evaluations.
The COVID-19 disease continues to spread around the whole world. Medical imaging and corresponding artificial intelligence applications together with clinical indicators provides solutions for triage, risk analysis and temporal analysis. This study provides a solution from multi-modal perspective to leverage the CT information but to inference on X-ray to avoid the necessity of taking CT imaging because of limited accessibility, dose and decontamination concern. Future work focuses on leveraging paired CT information to estimate severity and other higher dimensional measures. We believe that our study introduces a new trend of combining multi-modal training and single-modality inference. Although this study tries to leverage CT information as much as possible to aid the data-driven AI solution on X-rays, it should be noted that the extraction of relevant information is limited by the nature of X-ray imaging because of 2D projection as well as impaired visibility with the presence of the non-lung thick tissue. In addition different vendors may apply varying post-processing algorithms to suppress information on those thick tissue regions. To avoid information loss, our AI may be deployed directly on X-ray hardware with direct access to X-ray raw images for ideal translation to the clinic.
Compliance with Ethical Standards
The principles outlined in the Helsinki Declaration of 1975, as revised in 2000 are followed.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This study is internally funded by GE healthcare.
Biographies
Tao Tan is a GE employee in Netherlands
Bipul Das is a GE employee in India
Ravi Soni is a GE employee in US
Mate Fejes is a GE employee in Hungary
Hongxu Yang is a GE employee in Netherlands
Sohan Ranjan is a GE employee in India
Daniel Attila Szabo is a GE employee in Hungary
Vikram Melapudi is a GE employee in India
K S Shriram is a GE employee in India
Utkarsh Agrawal is a GE employee in India
Laszlo Rusko, is a GE employee in Hungary
Zita Herczeg is a GE employee in Hungary
Barbara Darazs is a GE employee in Hungary
Pal Tegzes is a GE employee in Hungary
Lehel Ferenczi is a GE employee in Hungary
Rakesh Mullick is a GE employee in India
Gopal Avinash is a GE employee in US
Communicated by Zidong Wang
References
- 1.Al-Awadhi Abdullah M., Al-Saifi Khaled, Al-Awadhi Ahmad, Alhamadi Salah. Death and contagious infectious diseases: Impact of the covid-19 virus on stock market returns. Journal of Behavioral and Experimental Finance. 2020 doi: 10.1016/j.jbef.2020.100326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marcus S Shaker, John Oppenheimer, Mitchell Grayson, David Stukus, Nicholas Hartog, Elena WY Hsieh, Nicholas Rider, Cullen M Dutmer, Timothy K Vander Leek, Harold Kim, et al., Covid-19: pandemic contingency planning for the allergy and immunology clinic, The Journal of Allergy and Clinical Immunology. In Practice, 2020. [DOI] [PMC free article] [PubMed]
- 3.Leung Kathy, Wu Joseph T., Liu Di, Leung Gabriel M. First-wave covid-19 transmissibility and severity in china outside hubei after control measures, and second-wave scenario planning: a modelling impact assessment. The Lancet. 2020 doi: 10.1016/S0140-6736(20)30746-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ali Inayat. Covid-19: Are we ready for the second wave? Disaster Medicine and Public Health Preparedness. 2020:1–3. doi: 10.1017/dmp.2020.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alberto Aleta, David Martin-Corral, Ana Pastore y Piontti, Marco Ajelli, Maria Litvinova, Matteo Chinazzi, Natalie E Dean, M Elizabeth Halloran, Ira M Longini Jr, Stefano Merler, et al., Modeling the impact of social distancing, testing, contact tracing and household quarantine on second-wave scenarios of the covid-19 epidemic, medRxiv, 2020. [DOI] [PMC free article] [PubMed]
- 6.Evenett Simon J., Winters L. Alan. Preparing for a second wave of covid-19 a trade bargain to secure supplies of medical goods. Global Trade Alert. 2020 [Google Scholar]
- 7.Jhu JHU, statistics about covid19 by john hopkins university, https://coronavirus.jhu.edu, 2020, Accessed: 2020-07-29.
- 8.Baud David, Qi Xiaolong, Nielsen-Saines Karin, Musso Didier, Pomar Léo, Favre Guillaume. Real estimates of mortality following covid-19 infection. The Lancet infectious diseases. 2020 doi: 10.1016/S1473-3099(20)30195-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimple D Rajgor, Meng Har Lee, Sophia Archuleta, Natasha Bagdasarian, and Swee Chye Quek, The many estimates of the covid-19 case fatality rate, The Lancet Infectious Diseases, vol. 20, no. 7, pp. 776–777, 2020. [DOI] [PMC free article] [PubMed]
- 10.Sung-mok Jung, Andrei R Akhmetzhanov, Katsuma Hayashi, Natalie M Linton, Yichi Yang, Baoyin Yuan, Tetsuro Kobayashi, Ryo Kinoshita, and Hiroshi Nishiura, Real-time estimation of the risk of death from novel coronavirus (covid-19) infection: inference using exported cases, Journal of clinical medicine, vol. 9, no. 2, pp. 523, 2020. [DOI] [PMC free article] [PubMed]
- 11.Liu Kai, Chen Ying, Lin Ruzheng, Han Kunyuan. Clinical features of covid-19 in elderly patients: A comparison with young and middle-aged patients. Journal of Infection. 2020 doi: 10.1016/j.jinf.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huaiyu Tian, Yonghong Liu, Yidan Li, Chieh-Hsi Wu, Bin Chen, Moritz UG Kraemer, Bingying Li, Jun Cai, Bo Xu, Qiqi Yang, et al., An investigation of transmission control measures during the first 50 days of the covid-19 epidemic in china, Science, vol. 368, no. 6491, pp. 638–642, 2020. [DOI] [PMC free article] [PubMed]
- 13.Gatto Marino, Bertuzzo Enrico, Mari Lorenzo, Miccoli Stefano, Carraro Luca, Casagrandi Renato, Rinaldo Andrea. Spread and dynamics of the covid-19 epidemic in italy: Effects of emergency containment measures. Proceedings of the National Academy of Sciences. 2020;117(19):10484–10491. doi: 10.1073/pnas.2004978117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lanciotti Robert S., Calisher Charles H., Gubler Duane J., Chang Gwong-Jen, Vance Vorndam A. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. Journal of Clinical Microbiology. 1992;30(3):545–551. doi: 10.1128/jcm.30.3.545-551.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hyungjin Kim, Hyunsook Hong, and Soon Ho Yoon, Diagnostic performance of ct and reverse transcriptase-polymerase chain reaction for coronavirus disease 2019: a meta-analysis, Radiology, p. 201343, 2020. [DOI] [PMC free article] [PubMed]
- 16.Kucirka Lauren M., Lauer Stephen A., Laeyendecker Oliver, Boon Denali, Lessler Justin. Variation in false-negative rate of reverse transcriptase polymerase chain reaction–based sars-cov-2 tests by time since exposure. Annals of Internal Medicine. 2020 doi: 10.7326/M20-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yicheng Fang, Huangqi Zhang, Jicheng Xie, Minjie Lin, Lingjun Ying, Peipei Pang, and Wenbin Ji, Sensitivity of chest ct for covid-19: comparison to rt-pcr, Radiology, p. 200432, 2020. [DOI] [PMC free article] [PubMed]
- 18.Tao Ai, Zhenlu Yang, Hongyan Hou, Chenao Zhan, Chong Chen, Wenzhi Lv, Qian Tao, Ziyong Sun, and Liming Xia, Correlation of chest ct and rt-pcr testing in coronavirus disease 2019 (covid-19) in china: a report of 1014 cases, Radiology, p. 200642, 2020. [DOI] [PMC free article] [PubMed]
- 19.Buddhisha Udugama, Pranav Kadhiresan, Hannah N Kozlowski, Ayden Malekjahani, Matthew Osborne, Vanessa YC Li, Hongmin Chen, Samira Mubareka, Jonathan B Gubbay, and Warren CW Chan, Diagnosing covid-19: the disease and tools for detection, ACS nano, vol. 14, no. 4, pp. 3822–3835, 2020. [DOI] [PubMed]
- 20.Thierry Blanchon, Jeanne-Marie Bréchot, Philippe A Grenier, Gilbert R Ferretti, Etienne Lemarié, Bernard Milleron, Dominique Chagué, François Laurent, Yves Martinet, Catherine Beigelman-Aubry, et al., Baseline results of the depiscan study: a french randomized pilot trial of lung cancer screening comparing low dose ct scan (ldct) and chest x-ray (cxr), Lung cancer, vol. 58, no. 1, pp. 50–58, 2007. [DOI] [PubMed]
- 21.Jacobi Adam, Chung Michael, Bernheim Adam, Eber Corey. Portable chest x-ray in coronavirus disease-19 (covid-19): A pictorial review. Clinical Imaging. 2020 doi: 10.1016/j.clinimag.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodolfo M Pereira, Diego Bertolini, Lucas O Teixeira, Carlos N Silla Jr, and Yandre MG Costa, Covid-19 identification in chest x-ray images on flat and hierarchical classification scenarios, Computer Methods and Programs in Biomedicine, p. 105532, 2020. [DOI] [PMC free article] [PubMed]
- 23.Isabella Castiglioni, Davide Ippolito, Matteo Interlenghi, Caterina Beatrice Monti, Christian Salvatore, Simone Schiaffino, Annalisa Polidori, Davide Gandola, Cristina Messa, and Francesco Sardanelli, Artificial intelligence applied on chest x-ray can aid in the diagnosis of covid-19 infection: a first experience from lombardy, italy, medRxiv, 2020. [DOI] [PMC free article] [PubMed]
- 24.Keelin Murphy, Henk Smits, Arnoud JG Knoops, Mike BJM Korst, Tijs Samson, Ernst T Scholten, Steven Schalekamp, Cornelia M Schaefer-Prokop, Rick HHM Philipsen, Annet Meijers, et al., Covid-19 on the chest radiograph: A multi-reader evaluation of an ai system, Radiology, p. 201874, 2020. [DOI] [PMC free article] [PubMed]
- 25.Feng Shi, Jun Wang, Jun Shi, Ziyan Wu, Qian Wang, Zhenyu Tang, Kelei He, Yinghuan Shi, and Dinggang Shen, Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for covid-19, IEEE reviews in biomedical engineering, 2020. [DOI] [PubMed]
- 26.Emanuele Neri, Vittorio Miele, Francesca Coppola, and Roberto Grassi, Use of ct and artificial intelligence in suspected or covid-19 positive patients: statement of the italian society of medical and interventional radiology, La radiologia medica, p. 1, 2020. [DOI] [PMC free article] [PubMed]
- 27.Jiang Dongsheng, Dou Weiqiang, Vosters Luc, Xiayu Xu., Sun Yue, Tan Tao. Denoising of 3d magnetic resonance images with multi-channel residual learning of convolutional neural network. Japanese Journal of Radiology. 2018;36(9):566–574. doi: 10.1007/s11604-018-0758-8. [DOI] [PubMed] [Google Scholar]
- 28.Kozegar Ehsan, Soryani Mohsen, Behnam Hamid, Salamati Masoumeh, Tan Tao. Mass segmentation in automated 3-d breast ultrasound using adaptive region growing and supervised edge-based deformable model. IEEE Transactions on Medical Imaging. 2017;37(4):918–928. doi: 10.1109/TMI.2017.2787685. [DOI] [PubMed] [Google Scholar]
- 29.Tan Tao, Platel Bram, Mus Roel, Tabar Laszlo, Mann Ritse M., Karssemeijer Nico. Computer-aided detection of cancer in automated 3-d breast ultrasound. IEEE Transactions on Medical Imaging. 2013;32(9):1698–1706. doi: 10.1109/TMI.2013.2263389. [DOI] [PubMed] [Google Scholar]
- 30.Tan Tao, Platel Bram, Huisman Henkjan, Sánchez Clara I., Mus Roel, Karssemeijer Nico. Computer-aided lesion diagnosis in automated 3-d breast ultrasound using coronal spiculation. IEEE Transactions on Medical Imaging. 2012;31(5):1034–1042. doi: 10.1109/TMI.2012.2184549. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Jiakang, Pan Fenxin, Li Wei, Hangtong Hu., Wang Wei, Huang Qinghua. Feature fusion for diagnosis of atypical hepatocellular carcinoma in contrast-enhanced ultrasound. IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control. 2021 doi: 10.1109/TUFFC.2021.3110590. [DOI] [PubMed] [Google Scholar]
- 32.Feng Xiangfei, Huang Qinghua, Li Xuelong. Ultrasound image de-speckling by a hybrid deep network with transferred filtering and structural prior. Neurocomputing. 2020;414:346–355. [Google Scholar]
- 33.Huang Qinghua, Huang Yonghao, Luo Yaozhong, Yuan Feiniu, Li Xuelong. Segmentation of breast ultrasound image with semantic classification of superpixels. Medical Image Analysis. 2020;61 doi: 10.1016/j.media.2020.101657. [DOI] [PubMed] [Google Scholar]
- 34.Huang Qinghua, Chen Yongdong, Liu Longzhong, Tao Dacheng, Li Xuelong. On combining biclustering mining and adaboost for breast tumor classification. IEEE Transactions on Knowledge and Data Engineering. 2019;32(4):728–738. [Google Scholar]
- 35.Huang Qinghua, Pan Fengxin, Li Wei, Yuan Feiniu, Hangtong Hu., Huang Jinhua, Jie Yu., Wang Wei. Differential diagnosis of atypical hepatocellular carcinoma in contrast-enhanced ultrasound using spatio-temporal diagnostic semantics. IEEE Journal of Biomedical and Health Informatics. 2020;24(10):2860–2869. doi: 10.1109/JBHI.2020.2977937. [DOI] [PubMed] [Google Scholar]
- 36.Annika Hänsch, Jan H Moltz, Benjamin Geisler, Christiane Engel, Jan Klein, Angelo Genghi, Jan Schreier, Tomasz Morgas, and Benjamin Haas, Hippocampus segmentation in ct using deep learning: impact of mr versus ct-based training contours, Journal of Medical Imaging, vol. 7, no. 6, pp. 064001, 2020. [DOI] [PMC free article] [PubMed]
- 37.Vignav Ramesh, Blaine Rister, and Daniel L Rubin, Covid-19 lung lesion segmentation using a sparsely supervised mask r-cnn on chest x-rays automatically computed from volumetric cts, arXiv preprint arXiv:2105.08147, 2021.
- 38.Eduardo Mortani Barbosa Jr. au2, Warren B. Gefter, Rochelle Yang, Florin C. Ghesu, Siqi Liu, Boris Mailhe, Awais Mansoor, Sasa Grbic, Sebastian Piat, Guillaume Chabin, Vishwanath R S., Abishek Balachandran, Sebastian Vogt, Valentin Ziebandt, Steffen Kappler, and Dorin Comaniciu, Automated detection and quantification of covid-19 airspace disease on chest radiographs: A novel approach achieving radiologist-level performance using a cnn trained on digital reconstructed radiographs (drrs) from ct-based ground-truth, 2020.
- 39.Tao Tan, Bipul Das, Ravi Soni, Mate Fejes, Sohan Ranjan, Daniel Attila Szabo, Vikram Melapudi, KS Shriram, Utkarsh Agrawal, Laszlo Rusko, et al., Pristine annotations-based multi-modal trained artificial intelligence solution to triage chest x-ray for covid-19, in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2021, pp. 325–334.
- 40.Wim van Aarle, Willem Jan Palenstijn, Jeroen Cant, Eline Janssens, Folkert Bleichrodt, Andrei Dabravolski, Jan De Beenhouwer, K. Joost Batenburg, and Jan Sijbers, Fast and flexible x-ray tomography using the astra toolbox, Opt. Express, vol. 24, pp. 25129–25147, Oct 2016. [DOI] [PubMed]
- 41.Olaf Ronneberger, Philipp Fischer, and Thomas Brox, U-net: Convolutional networks for biomedical image segmentation, 2015.
- 42.Diederik P. Kingma and Jimmy Ba, Adam: A method for stochastic optimization, 2017.
- 43.Pneumonia Kaggle, Kaggle rsna pneumonia, https://www.kaggle.com/c/rsna-pneumonia-detection-challenge, 2018, Accessed: 2020-06-30.
- 44.Chest Kaggle, Kaggle chest, https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia, 2017, Accessed: 2020-06-30.
- 45.Aurelia Bustos, Antonio Pertusa, Jose-Maria Salinas, and Maria de la Iglesia-Vayá, Padchest: A large chest x-ray image dataset with multi-label annotated reports, arXiv preprint arXiv:1901.07441, 2019. [DOI] [PubMed]
- 46.Github IEEE, Covid-19 image data collection, https://github.com/ieee8023/covid-chestxray-dataset/, 2020, Accessed: 2020-06-30.
- 47.Wang Xiaosong, Peng Yifan, Le Lu., Zhiyong Lu., Bagheri Mohammadhadi, Summers Ronald M. Proceedings of the IEEE conference on computer vision and pattern recognition. 2017. Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases; pp. 2097–2106. [Google Scholar]
- 48.Linda Wang, Zhong Qiu Lin, and Alexander Wong, Covid-net: a tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images, Scientific Reports, vol. 10, no. 1, pp. 19549, Nov 2020. [DOI] [PMC free article] [PubMed]
- 49.Maria de la Iglesia Vayá, Jose Manuel Saborit-Torres, Joaquim Angel Montell Serrano, Elena Oliver-Garcia, Antonio Pertusa, Aurelia Bustos, Miguel Cazorla, Joaquin Galant, Xavier Barber, Domingo Orozco-Beltrán, Francisco García-García, Marisa Caparrós, Germán González, and Jose María Salinas, Bimcv covid-19+: a large annotated dataset of rx and ct images from covid-19 patients, 2021.
- 50.Muhammad EH Chowdhury, Tawsifur Rahman, Amith Khandakar, Rashid Mazhar, Muhammad Abdul Kadir, Zaid Bin Mahbub, Khandakar Reajul Islam, Muhammad Salman Khan, Atif Iqbal, Nasser Al Emadi, et al., Can ai help in screening viral and covid-19 pneumonia?, IEEE Access, vol. 8, pp. 132665–132676, 2020.
- 51.Jeremy Irvin, Pranav Rajpurkar, Michael Ko, Yifan Yu, Silviana Ciurea-Ilcus, Chris Chute, Henrik Marklund, Behzad Haghgoo, Robyn Ball, Katie Shpanskaya, et al., Chexpert: A large chest radiograph dataset with uncertainty labels and expert comparison, in Proceedings of the AAAI conference on artificial intelligence, 2019, vol. 33, pp. 590–597.
- 52.Rodriguez-Ruiz Alejandro, Krupinski Elizabeth, Mordang Jan-Jurre, Schilling Kathy, Heywang-Köbrunner Sylvia H., Sechopoulos Ioannis, Mann Ritse M. Detection of breast cancer with mammography: effect of an artificial intelligence support system. Radiology. 2019;290(2):305–314. doi: 10.1148/radiol.2018181371. [DOI] [PubMed] [Google Scholar]
- 53.van Zelst J.C.M., Tan T., Platel B., de Jong M., Steenbakkers A., Mourits M., Grivegnee A., Borelli C., Karssemeijer N., Mann R.M. Improved cancer detection in automated breast ultrasound by radiologists using computer aided detection. European Journal of Radiology. 2017;89:54–59. doi: 10.1016/j.ejrad.2017.01.021. [DOI] [PubMed] [Google Scholar]
- 54.DeGrave Alex J., Janizek Joseph D., Lee Su-In. Ai for radiographic covid-19 detection selects shortcuts over signal. Nature Machine Intelligence. 2021:1–10. [Google Scholar]
- 55.Xueyan Mei, Hao-Chih Lee, Kai-yue Diao, Mingqian Huang, Bin Lin, Chenyu Liu, Zongyu Xie, Yixuan Ma, Philip M Robson, Michael Chung, et al., Artificial intelligence–enabled rapid diagnosis of patients with covid-19, Nature Medicine, pp. 1–5, 2020. [DOI] [PMC free article] [PubMed]
- 56.WHO, Use of chest imaging in COVID-19, A RAPID ADVICE GUIDE, WHO, 2020. [DOI] [PMC free article] [PubMed]