Preprint
Article
EXtra-Xwiz: A Tool to Streamline Serial Femtosecond Crystallography Workflows at European XFEL
Altmetrics
Downloads
164
Views
117
Comments
0
A peer-reviewed article of this preprint also exists.
This version is not peer-reviewed
Submitted:
18 September 2023
Posted:
21 September 2023
You are already at the latest version
Alerts
Abstract
X-ray free electron lasers deliver photon pulses that are bright enough to observe diffraction from extremely small crystals, at a time scale outrunning their destruction. As crystals are continuously replaced, this technique is termed serial femtosecond crystallography (SFX). Due to its high pulse repetition rate, the European XFEL enables the collection of rich and extensive data sets, suited to study various scientific problems including ultrafast processes. The enormous data rate, data complexity, and the nature of the pixelized multi-modular area detectors at European XFEL pose severe challenges to users. To streamline the analysis of SFX data, we developed the semi-automated pipeline EXtra-Xwiz around the established CrystFEL program suite, processing diffraction patterns on detector frames into structure factors. Here we present EXtra-Xwiz, and introduce its architecture and use by means of a tutorial. Future plans for its development and expansion are also discussed.
Keywords:
Subject: Computer Science and Mathematics - Software
1. Introduction
The structural arrangement of atoms in matter and the nature of their chemical bonds can be deciphered by employing X-ray crystallography. This experimental technique has been pivotal in structural biology (see, e.g., [1,2,3] and references therein) and contributes to date to approximately 85% of the structures released in the Protein Data Bank (calculated using data from [4,5]). X-ray crystallography requires samples to be present in the form of crystals: periodic repetitions of a unique unit cell which results in Bragg peaks upon exposure to X-rays. These encode part of the information to reconstruct the electron density of the sample. In a simplified description, assuming that the kinematic approximation holds, the overall intensity of Bragg peaks increases with (i) the squared number of unit cells in the crystal and its degree of perfection, and (ii) the number of photons interacting with the sample 1. The size and quality of crystals can only be controlled to a limited extent, also due to the need to maintain realistic near-physiological conditions when biological compounds are investigated. The key to elucidate increasingly complex macromolecules, from e.g., Hemoglobin [7] to the Ribosome [8], instrumental to advance structural biology, has thus been the exploitation of more and more advanced photon sources. The photon flux generated from modern sources, which are either based on storage rings or X-ray free electron lasers (XFELs), has enabled the collection of diffraction data allowing up to Ångstrom resolution from smaller and smaller crystals, down to sub-micrometer size at XFELs as a result of their exceptional brilliance. The interaction of crystals with intense X-ray beams results in several physical processes that can lead to permanent radiation damage (see, e.g., [9]). Classical crystallography experiments consist of rotation scans exposing repeatedly the same region of the crystal. Consequently, it accumulates damage during data acquisition. To mitigate this, cryogenic conditions can be employed so as to hamper the processes of radical formation from photolectrons [10]. Another strategy consists in collecting data from fresh regions of the sample, either of the same crystal or different ones, in a serial fashion (see, e.g., [11]). The latter paradigm is adopted at XFELs as a single X-ray pulse typically deposits enough energy to completely destroy the sample [12]. However, the temporal duration of pulses is short enough – of the order of several femtoseconds – that signals from almost undamaged crystals can be detected, as the timescale of the ion movement is much longer 2. At XFELs, crystals are continuously replaced typically using liquid jets or movable fixed-target stages and intercept the X-ray beam with a certain probability (the so-called hit rate). This technique is termed serial femtosecond crystallography (SFX) [14,15,16,17,18]. Furthermore, owing to the femtosecond duration of their pulses, XFELs are exceptional tools to perform time-resolved investigations at resolutions not achievable by photon sources based on storage rings [13,19,20,21,22,23].
In parallel to the development of X-ray sources and instrumentation, the computer hardware and software to process data from raw diffraction images to the final structural model has evolved in several aspects, including the development of novel crystallographic methods, the usage of parallel processing computational methods, and the design of graphical interfaces to facilitate and automate the data processing. The processing of X-ray crystallography data commences with the reduction of raw detector frames to a unique set of structure factors [24]. This includes finding Bragg peaks, indexing them, integrating pixel intensities in three-dimensions, and averaging the symmetry-equivalent reflection observations with proper scaling. These steps have been implemented in popular software packages such as XDS [25], Mosflm [26], or DIALS [27]. Owing to the nature of data collection, serial crystallography requires different algorithms and approaches, and includes a hit-finding step for identifying detector frames containing the signature of diffraction, which is a prerequisite for indexing the reciprocal lattice [17]. Also in this case, dedicated software suites have been developed in the last decade. Notably, DIALS has been extended to process serial crystallography data [28], and the CrystFEL suite [29] has been developed. Subsequent data analysis, from structure factors to the final atomic model, requires software for crystallographic phasing, model building from derived electron density and its refinement and validation 3.
The entire crystallography data processing pipeline consists of the sequential execution of several tasks, often performed by different software programs. As the input and output data formats, as well as the user experience, might differ greatly across tools, several software pipelines have been developed with the aim of abstracting complexity and increasing analysis throughput and automation level [30,31,32,33]. In fact, X-ray crystallography beamlines at storage rings are exceptional examples of sophisticated ecosystems including state-of-the-art robotics, information systems and processing tools, allowing for a comprehensive automation [30,34,35]. Such simplification empowers inexperienced users to focus on scientific questions. It should be pointed out that the need for expert knowledge persists in demanding cases, e.g., when the diffraction signal is particularly weak, or extensive parameter optimization is required.
Several challenges are intrinsic to serial crystallography at XFELs, such as pulse-to-pulse jitter of the X-ray beam in space, wavelength, and energy, which are typically reflected in the amount of diagnostics necessary to interpret the outcome of the experiment. Additionally, the rate and amount of data collected to solve the scientific problem under investigation as well as the often-complicated nature of custom-built detectors further complicate processing and interpretation [15,17,36]. For example, the European XFEL (EuXFEL) [37,38] generates up to 27,000 X-ray pulses per second. A fraction of these is collected by multi-modular pixelized area detectors, such as the Adaptive-Gain Integrating Pixel Detector (AGIPD, up to 3,520 frames or 14 GiB per second) [39], the Large Pixel Detector (LPD, up to 5,120 frames or 10 GiB per second) [40], and the JUNGFRAU detector (up to 160 frames or 1.9 GiB per second) [41], which are synchronized with X-ray pulses. Due to technical reasons, the data acquisition system stores each detector module separately. In particular, predefined sequences of data from each module are stored in different HDF5 4 files in the EuXFEL data format (EXDF), which might be a significant barrier for several users, and currently cannot be used directly by popular software like CrystFEL. Additionally, the sheer volume of the data to be analysed makes workflows practically unfeasible unless distributed computing on high-performance computing (HPC) clusters is employed, whose usage is an additional burden to scientists. Finally, photon sources like EuXFEL enable investigation of ultrafast processes, which are often performed utilizing some form of excitation of the sample (the so-called pump) to then probe the induced molecular dynamics with the XFEL beam. The cost of this is tedious bookkeeping of the data frame subsets, given the pump-probe patterns and verification of correct time sampling, and the same applies in general to diagnostic means.
With the aim of abstracting as much complexity as possible so as to allow scientists to focus on their biological question, we developed EXtra-Xwiz [43]. This allows for a high degree of automation of data analysis workflows through its integration with other services provided at EuXFEL. In this paper, we introduce EXtra-Xwiz and discuss its current status and future goals. In Section 2 we describe the EXtra-Xwiz design and architecture, followed by a step-by-step tutorial with an example of processing SFX data in Section 3. Finally, we give an outlook on planned extensions to the pipeline in Section 4.
2. Design and implementation of the EXtra-Xwiz pipeline
EXtra-Xwiz consists of an SFX workflow-managing tool bundled with some auxiliary programs. In the simplest scenario, a workflow is run as a linear pipeline that will (i) prepare input data, (ii) distribute the input frames into subsets that can be processed in parallel, (iii) perform merging and scaling of structure factor intensity observations, and (iv) create crystallographic figures of merit (FOMs) [44].
To accomplish this, EXtra-Xwiz mostly utilizes programs from the CrystFEL suite. These include:
- indexamajig – main command-line program for indexing and integrating diffraction patterns;
- cell_explorer – a tool for displaying and determining unit cell parameters of the crystalline sample;
- partialator – for scaling, merging, and post-refining reflections data;
- check_hkl – for calculating FOMs based on the full set of merged reflections, such as completeness, average signal strengths, and redundancy;
- compare_hkl – for calculating FOMs based on the merged reflections split into two sets, such as R-factors and correlation coefficients.
The full list of the CrystFEL programs can be found in the dedicated publication [45] and documentation [46]. CrystFEL is freely available at [46].
EXtra-Xwiz is released as an open source project at [47].
2.1. Structure of the EXtra-Xwiz pipeline
A schematic representation of the pipeline is shown in Figure 1. EXtra-Xwiz requires a configuration file with parameters for each step, organized into sections.
The first step of EXtra-Xwiz pipeline is to provide the data in a format suitable for processing by CrystFEL. CrystFEL can read diffraction data stored in Crystallographic Binary Format (CBF) or Hierarchical Data Format v.5 (HDF5) [48]. For the latter, various standards are supported, including NeXus [49,50] and the native format of the Coherent X-ray Imaging Data Bank (CXIDB) [51].
Due to technical requirements, data from each module of X-ray detectors at EuXFEL is saved to separate files. As this is not compatible with CrystFEL, the library EXtra-data [52] is used to generate a single HDF5 file with a "virtual" layout (virtual dataset file, VDS) containing references to the relevant original EXDF data. This processing step, as illustrated in Figure 2, is represented as a block "Data" in Figure 1 and corresponds to the section "[data]" in the configuration file.
To cope with variable spatial configurations of detector modules, CrystFEL uses a generalized detector representation – the geometry file [29]. This describes the position of each detector pixel in the laboratory coordinate system, as well as the pixel size and the X-ray beam energy. It also contains information on the internal HDF5 paths of the image dataset and any bad pixels mask in the input data file. The geometry file, represented with the block "Geometry" in Figure 1, is specified in the section "[geom]" of the EXtra-Xwiz configuration file.
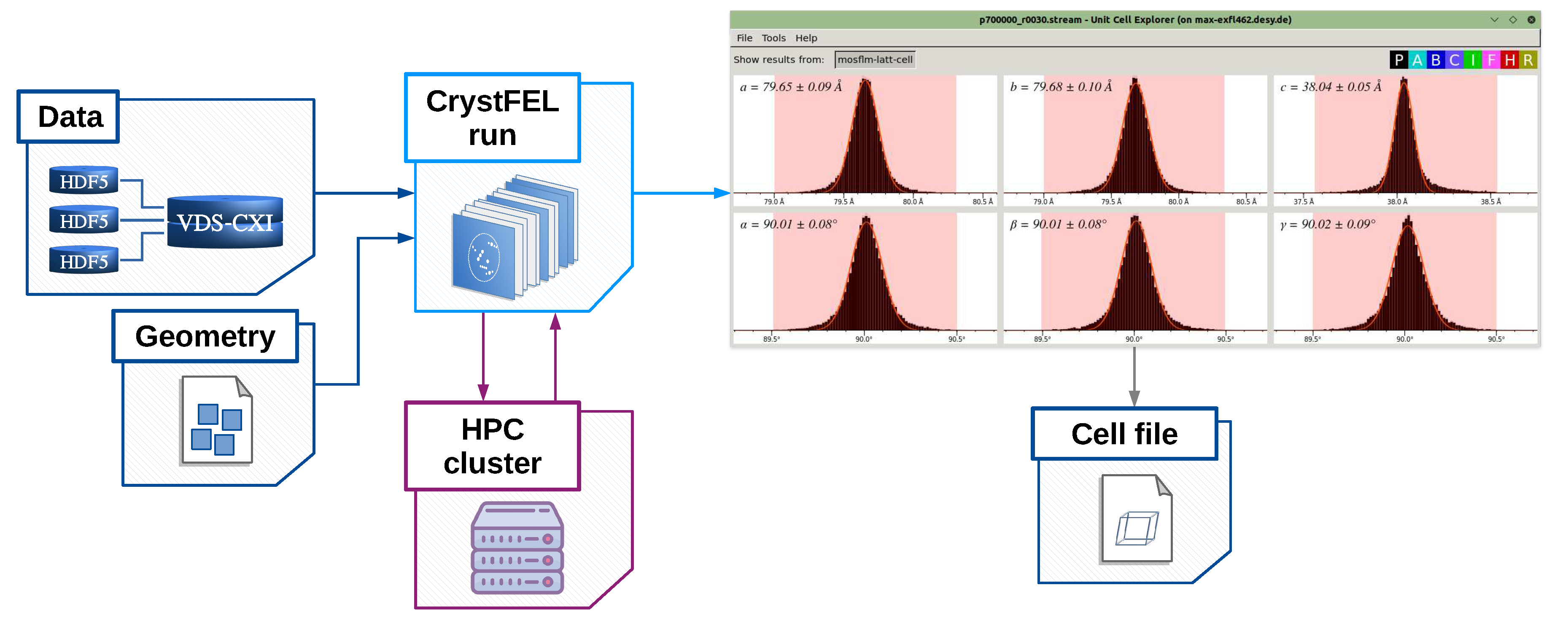
If prior information on the unit cell is known, it can be used to filter auto-indexing results that agree to the expected reference. Such reference unit cell parameters (a, b, c, , , ) can be specified in the unit cell file schematically represented with a block "Unit cell" in Figure 1. It can be provided to EXtra-Xwiz in the "[unit_cell]" section of the configuration file. Some of the indexing methods can derive the crystal symmetry even without prior information on the lattice. In this case, EXtra-Xwiz can be run without specifying the unit cell file, as illustrated in Figure 3. After the indexing step, the cell_explorer graphical user interface (GUI) will be launched to display the histograms of resulting lattice parameters. This allows users to determine the cell parameters with a fit and to save them into a unit cell file which will be expected by EXtra-Xwiz to continue processing the data as shown in Figure 1.
Main processing and reduction of the SFX data in EXtra-Xwiz is performed with the indexamajig tool from CrystFEL. It is used for finding the location of Bragg peak candidates in each detector image, indexing the patterns, and integrating peak intensities. For each image frame CrystFEL writes information on identified Bragg peak candidates, lattice parameters found by the auto-indexing algorithm, and integrated intensities into a so-called stream file in a form of a plain text. This processing step is represented with a "CrystFEL run" block in Figure 1. Parameters for indexamajig should be specified in the "[indexamajig_run]" section of the EXtra-Xwiz configuration file.
Identifying optimal parameters for the Bragg peaks search is crucial for successful indexing. If too many true peaks are missed or too many false peaks (for example, from the detector background or misbehaving detector pixels) are included, the indexing algorithm may fail completely or identify wrong unit-cell parameters and crystal orientation. The sample and data taking conditions may differ significantly during the experiment and therefore it is a good practice to regularly tune parameters. The graphical user interface of the CrystFEL suite offers a convenient tool for performing such a preparative step based on visual inspection of a few of image frames. A session of the CrystFEL GUI can be stored into a project file and EXtra-Xwiz provides a command-line tool xwiz-import-project for importing parameters from such file into the EXtra-Xwiz configuration file. This procedure is represented by the "CrystFEL GUI proj." block in Figure 1.
The most time-consuming step of processing SFX data is indexing. Due to the independent nature of the SFX frames, portions of collected data sets can be processed separately, which allows for parallelization. EXtra-Xwiz automatically distributes indexamajig jobs on the local HPC cluster Maxwell operated with the Slurm [53] batch-queue system. The results are then concatenated into a single stream file. This is represented by the "HPC cluster" block in Figure 1. In the configuration file the name of the Slurm partition as well as the number and expected duration of distributed tasks have to be specified under the "[slurm]" section.
After indexing, reflection intensities from the whole data set are merged for each symmetrically unique reflection. This task is accomplished by the CrystFEL partialator program, which can determine scaling factors for individual measurements, correct for partiality, and post-refine the model parameters for each crystal [54]. This process is usually repeated in an iterative procedure to bring the individual measurements into agreement [48,54]. The partialator program operates on the content of the stream file and therefore does not require the initial SFX data. This data analysis step corresponds to the "partialator run" block in Figure 1 and "[merging]" section of the EXtra-Xwiz configuration file. Merged reflections are stored in the hkl file format as plain text, which can be imported into most structure-solution packages. Output from partialator is used by EXtra-Xwiz to calculate figures of merit with check_hkl and compare_hkl. The completeness of unique reflections and signal-to-noise ratio (SNR) are estimated with check_hkl from a single list of reflections merged from all available data. To calculate the correlation coefficients and as well as R-factor , the reflection observations for each unique Miller index are randomly split before merging by partialator into two halves. Both half-data sets are merged separately and their correlation computed with compare_hkl. Values of the listed FOMs for all data and outer reflections shell are stored by EXtra-Xwiz into a table in a summary file.
Certain types of SFX experiments are performed varying some external parameter, which results in data subsets having different characteristics. These include time-resolved SFX experiments, in which sample states excited for example by visible light (pump on) are probed by X-rays as a function of time. Interpretation of such data usually relies on the difference between the subsets. It is therefore crucial to ensure that all other experimental conditions, e.g., the X-ray beam fluence, as well as data processing are kept as close as possible between the sets. To satisfy this, pump on frames are often collected interleaved with the pump off frames within the same train of X-ray pulses. In this case, partialator can be used to split reflections after scaling and post-refinement and prior to merging. This option requires a text file with a "data set identifier" (e.g., the pump status) for each frame in the input data. EXtra-Xwiz produces such files either using the frame pattern information specified by a user, or automatically utilizing information from a diode which records the signal from the pump laser. This procedure is represented with the "partialator split" block in Figure 1. Parameters for the splitting have to be specified in an optional "[partialator_split]" section of the EXtra-Xwiz configuration file. Merged reflections hkl files as well as FOMs are produced for each subset as well as for the overall data set.
The execution of EXtra-Xwiz can be automatically triggered as soon as experimental data become available by exploiting its integration with the software DAMNIT [55]. Furthermore, DAMNIT provides a graphical interface to EXtra-Xwiz results.
3. Data processing with EXtra-Xwiz by example
This section describes an example of processing SFX data from hen egg-white lysozyme (HEWL) microcrystals collected at the SPB/SFX instrument using the AGIPD detector (run 30, proposal 700000). Only basic knowledge of the Unix commands and environment are expected from the reader. Lines starting with a "$" indicate commands which should be executed in a Unix shell. For readers who are not users of the European XFEL, the Virtual Infrastructure for Scientific Analysis (VISA) [56] service can be used and additional instructions are provided in Section 3.2.
Access to the Maxwell cluster is exclusive to EuXFEL users and detailed instructions on how to connect to it can be found in the EuXFEL Data Analysis user documentation [57]. In general, a user with an active account can connect to one of the interactive cluster nodes:
$ ssh <user name>@max-exfl-display.desy.de
To start using EXtra-Xwiz a dedicated module has to be loaded at the cluster with:
$ module load exfel EXtra-xwiz/crystals2023
As mentioned in Section 2.1, EXtra-Xwiz requires a configuration file in TOML format [58] for its operation 5. It should be named "xwiz_conf.toml" and a template of such file can be generated by starting the pipeline for the first time in an empty folder with the following command:
$ xwiz-workflow
The configuration file contains parameters for each of the pipeline processing steps organized into sections such as "[data]", "[geom]", "[unit_cell]", "[indexamajig_run]", and "[merging]". A copy of the configuration file used in this example along with all other files necessary for the pipeline execution can be downloaded from [47].
Data to be processed by the pipeline can be specified with just a proposal number and a list of runs in the "[data]" section of the configuration file:
[data] proposal = 700000 runs = [30]
It is possible to select a subset of frames from each run with an optional frames_range parameter in the same section, for example:
frames_range = {start = 0, end = 200000, step = 1}
This parameter has values organized into a dictionary similar to the Python range object but inclusive for the end value with end = -1 representing the last frame of the run.
For the purpose of reproducibility of the SFX analysis, EXtra-Xwiz supports a list of different versions of the CrystFEL suite which can be selected in the "[crystfel]" section:
[crystfel] version = '0.10.2'
Currently, recent major CrystFEL versions are available, as well as a "maxwell_dev" option which corresponds to the constantly updated installation of the latest CrystFEL version.
Geometry and unit cell parameters files should be provided to the pipeline in the "[geom]" and "[unit_cell]" sections, respectively:
[geom] file_path = "agipd_p700000_r0030.geom" [unit_cell] file_path = "hewl.cell"
A representative detector frame is shown in Figure 2(b). There detector modules are positioned in the laboratory frame layout with the use of EXtra-geom library [60].
In case the unit cell of the sample is not known prior to the analysis, it can be generated by setting the option file_path = "none" as described in Section 2.1. During the EXtra-Xwiz session, after the indexing step, an interactive cell_explorer session will start. At this point, the user is expected to determine the cell parameters from the histograms of indexing results (as explained in [48]), and save them into a unit cell file which will be requested by EXtra-Xwiz. This procedure is illustrated in Figure 3.
Parameters for Bragg peak identification and indexing with indexamajig program have to be specified in the "[indexamajig_run]" configuration block:
[indexamajig_run] resolution = 4.0 peak_method = "peakfinder8" peak_threshold = 800 peak_snr = 5 index_method = "mosflm" integration_radii = "2,3,5" ... min_peaks = 10 extra_options = "--no-non-hits-in-stream"
Documentation regarding all indexamajig options can be found at [46]. In the current state of EXtra-Xwiz not all of these options are covered by the default configuration file parameters, and if any of such options are required for data processing they can be specified in the string for extra_options parameter. Data processing with indexamajig is the most time-consuming step of the whole pipeline but the computations are usually performed in parallel on multiple nodes of the Maxwell cluster. Cluster partition, number of nodes to use in parallel and maximum expected duration of the individual jobs should be specified under "[slurm]" section of the configuration file:
[slurm] partition = "upex" n_nodes_all = 20 duration_all = "10:00:00"
For testing the pipeline on a small subset of data (e.g., a hundred frames), without exploiting the Slurm jobs scheduler, it is advised to select the "local" partition. In this case EXtra-Xwiz will run indexamajig on the same node the pipeline is running.
Reflection intensities obtained from the Bragg peaks indexing are merged and post-refined with the partialator tool and required parameters have to be specified under the "[merging]" block of EXtra-Xwiz configuration:
[merging] point_group = "422" scaling_model = "unity" scaling_iterations = 1 max_adu = 100000
Point groups corresponding to the symmetry groups of crystallized samples can be identified with the table in CrystFEL documentation [61].
In case of the time-resolved SFX experiments pump on (sample illuminated with the "pump" laser) and pump off (sample in the non-excited state) frames are processed in the same manner and separated only on the merging step of partialator. As already mentioned in Section 2.1, for such separation partialator requires an additional input file labelling accordingly each frame of the input data. EXtra-Xwiz can generate such file according to parameters specified in the "[partialator_split]" block. Let us assume that the machine delivers only at 1/8th of the 4.5 MHz maximum repetition rate and the detector is configured to record only these pulses. The sample is illuminated by infrared light every third delivered pulse (i.e., the 24th assuming 4.5 MHz operation), resulting in the following labels, "pump_on pump_off pump_off pump_on ...". The latter can be set in the configuration as:
[partialator_split]
execute = true
mode = "by_pulse_id"
[partialator_split.manual_datasets]
pump_on = {start=0, end=-1, step=24}
pump_off = [{start=8, step=24}, {start=16, step=24}]
Any user-defined set of labels can be specified with a corresponding list of inclusive range-like dictionary objects or pulse id values. Usually, in time-resolved experiments a diode is used to record data relative to the state of the pump laser. EXtra-Xwiz can utilize signal from this diode and automatically generate labels accordingly if the mode parameter is set to either "on_off" or "on_off_numbered":
[partialator_split] execute = true mode = "on_off_numbered" xray_signal = ["SPB_LAS_SYS/ADC/UTC1-1:channel_0.output", "data.rawData"] laser_signal = ["SPB_LAS_SYS/ADC/UTC1-1:channel_1.output", "data.rawData"]
The difference between the "on_off_numbered" and the "on_off" mode is that in the former case consequent events of the same kind (e.g., pump off) are identified by an increasing number. For the example given above, the "on_off_numbered" labels are "on_1 off_1 off_2 on_1, ...". Paths to the diode data specified for xray_signal and laser_signal are provided by beamline scientists. As data used in this tutorial does not originate from the pump-probe experiment, the splitting into datasets does not make sense and should be avoided by either setting execute = false or simply removing the "[partialator_split]" block from the configuration file.
After all the configuration parameters have been set the EXtra-Xwiz pipeline can be executed in an automatic mode with:
$ xwiz-workflow -a
Without the "-a" ("–automatic") optional argument the pipeline will verify each configuration parameter with a user in the interactive procedure. When the EXtra-Xwiz operation finishes, it will generate a summary file containing information on the processed data statistics and FOMs, for example:
Step # d_lim source N(crystals) N(frames) Indexing rate [%]
1 1.6 indexamajig 46899 639616 7.3
...
Crystallographic FOMs:
overall outer shell
Completeness 100.0 100.0
Signal-over-noise 4.224 0.99
CC_1/2 0.8974 0.03244
CC* 0.9726 0.2507
R_split 27.28 80.6
These results are provided only to demonstrate the capabilities of the pipeline and could be improved, for example by tuning selected parameters and processing more runs of the collected data. The resulting CrystFEL stream file can be found in the same folder and the output hkl file with full FOMs tables in the "partialator" folder.
3.1. Automatic scan over EXtra-Xwiz configuration parameters
Sometimes it is required to run the EXtra-Xwiz pipeline modifying one or multiple configuration parameters over a list of values, for example to assess the sensitivity of a given parameter. For such use case an xwiz-scan-parameters tool have been developed. Similar to xwiz-workflow, it requires a configuration file, and a template is generated on the first use of the tool in an empty folder:
$ xwiz-scan-parameters
The configuration file "xwiz_scan_conf.toml" for parameters scan tool consists of four sections: "[settings]", "[xwiz]", "[scan]", and "[output]". Main parameter of the "[settings]" block is xwiz_config which determines the path to the initial EXtra-Xwiz configuration file.
The "[scan]" section can contain any number of sub-sections. Each sub-section will be treated as a next level of a nested loop, therefore number of iterations in each scan are multiplied. Names of the parameters within each scan sub-section represent full names of the parameters in the EXtra-Xwiz configuration files and their values should contain either a list or an inclusive range-like dictionary of values to scan over. If multiple parameters are listed within one scan they have to contain the same number of values which will be modified simultaneously on each step of the scan, for example:
[scan.SNR]
'indexamajig_run.peak_snr' = {start = 3, end = 7, step = 2}
'indexamajig_run.peak_threshold' = [1000, 800, 700]
Here a scan with 3 iterations is defined: first an SNR value of 3 will be used with a threshold of 1000, next an SNR of 5 will be set with a threshold of 800, and finally an SNR of 7 with a threshold of 700.
In the "[output]" section, the file names to store the tables with the results can be specified. When the configuration file is ready, the parameters scan can be started by running the xwiz-scan-parameters in the same folder. It will run EXtra-Xwiz sequentially over all scan iterations and collect data processing statistics and overall values of FOMs for each scan step into a table similar to this one:
index_rate(%) ... cc_half cc_star r_split
peak_snr peak_threshold
3 1000 0.080 ... 0.051 0.313 105.40
5 800 7.332 ... 0.894 0.972 27.44
7 700 7.219 ... 0.919 0.979 25.81
3.2. Running EXtra-Xwiz tutorial at VISA
VISA is a platform for cloud-based data analysis, developed by the Institute Laue-Languevin and deployed at several European photon and neutron facilities [56]. Its documentation can be found at [62].
In order to perform this tutorial using VISA, an instance containing an EXtra-Xwiz installation can be generated and used in a web browser:
- navigate to https://visa.xfel.eu;
- click "Create a new instance";
- click "Search for experiments" and select the proposal "p700000 - SFX on Hen egg-white lysozyme, AGIPD detector";
- click on "EXtra-Xwiz_Crystals2023" environment;
- choose the virtual hardware;
- create the instance.
There is no need to load any additional modules and the tutorial described above applies except for the distribution of computations on the HPC cluster, which is not accessible from VISA. Because of that, the following should be used:
[slurm] partition = "local" [indexamajig_run] n_cores = 1 ...
Be careful, this would result in a very slow processing of the input data. Therefore, instead of running EXtra-Xwiz on the entire run, a pre-selection of "good frames" can be used by specifying in the "[data]" section:
[data] ... frames_list_file = "indexed_p700000_r0030.lst"
Please note that the "indexed_p700000_r0030.lst" file has been produced specifically for this VISA example and this option is never used in the actual processing of the experimental data.
4. Discussion and outlook
A challenge that requires expert knowledge and/or iterative processing steps lies in the optimization of parameters, such as minimum signal-to-noise ratio, peak finder thresholds, etcetera. Technically, as these parameters are passed to the CrystFEL programs through a configuration file, iterative runs require a re-editing of the configuration, which can quickly become tedious. Currently, EXtra-Xwiz offers two ways to simplify this: first, the software includes a grid search, as described in Section 3.1, that can scan over some parameter space. This requires some estimate of reasonable parameter ranges, and enough time to compute the necessary grid nodes. Second, peak-finding parameter optimization can be done manually, and with visual feedback, using the CrystFEL GUI. Results of a GUI session can be stored to a project file to be used by EXtra-Xwiz for batch processing of one or more entire runs. Both of these approaches have their limitations, in particular related to the interpretation of their results by inexperienced users. Alternatively to brute-force searches, optimization methods, for example based on artificial intelligence, can be employed. In particular, we developed a method based on Bayesian optimization that optimizes EXtra-Xwiz parameters by maximizing the indexing rate, and reduces the need for expertise in the interpretation of results [63]. This solution is being deployed at EuXFEL and integrated into EXtra-Xwiz. The same approach can also be used to tune the detector geometry representation in laboratory space. Indeed, the overall SFX workflow can greatly benefit from automating this task. At the moment, this is largely done manually. The operator is typically using graphical software for the visual centering and quadrant fitting of powder diffraction rings, followed by iterations of indexamajig and geoptimizer [64] for maximizing the yield of indexable frames.
As a more long-term outlook, we strive to give users an end-to-end experience, that is, a pipeline covering all the steps from detector images to an atomic structure built into the reconstructed electron density. For this purpose, downstream modules handling the program execution related to the tasks of crystallographic phasing, model building and (preliminary but automated) structure refinement need to be included to the EXtra-Xwiz framework. Furthermore, owing to the modular nature of EXtra-Xwiz, in the future, software different from CrystFEL (e.g., DIALS), might be considered. It should be pointed out that challenging problems, in particular with respect to de novo structures, will require user experience for decisions along the way, for instance which phasing method to apply, or which search model to use for molecular replacement. This means that the design of the workflows have to be well thought, allowing user intervention where required and good balance of parameters with reasonable defaults versus expert input, boiling down to a useful semi-automatic approach. For this, an improved user interface is being designed.
5. Conclusions
EXtra-Xwiz has been designed to support scientists performing SFX at the European XFEL. It handles tasks such as data preparation, discrimination of data based on pump status, and parallel processing which often pose a significant barrier to inexperienced users.
Acknowledgments
We appreciate the contributions to the construction, maintenance, and operation of the European XFEL and scientific instruments by the many people who are not listed as authors. We would like to thank all members of the EuXFEL Data Analysis group for continuous development and improvement of software tools and libraries for efficient data analysis. We are grateful to Philipp Schmidt for valuable feedback on the manuscript and his support. We would like to especially thank Thomas White, main developer of the CrystFEL suite, for the great software for the analysis of serial crystallography data and his invaluable input and feedback to our project. The authors would like to acknowledge Oleksandr Yefanov whose constructive criticism allowed us to shape a clear vision for the evolution of our framework. We would also like to express gratitude to the users of our software, who provided continuous feedback and useful suggestions.
Abbreviations
The following abbreviations are used in this manuscript:
| SFX | Serial femtosecond crystallography |
| XFEL | X-Ray Free-Electron Laser |
| EuXFEL | European XFEL facility |
| EXDF | EuXFEL Data Format |
| AGIPD | Adaptive-Gain Integrating Pixel Detector |
| LPD | Large Pixel Detector |
| HPC | High-Performance Computing |
| HDF5 | Hierarchical Data Format v.5 |
| CBF | Crystallographic Binary Format |
| CXIDB | Coherent X-ray Imaging Data Bank |
| GUI | Graphical User Interface |
| VDS | Virtual Dataset File |
| FOM | Figure Of Merit |
| SNR | Signal-to-Noise Ratio |
| HEWL | Hen Egg-White Lysozyme |
| VISA | Virtual Infrastructure for Scientific Analysis |
References
- Smyth, M.S.; Martin, J.H.J. x Ray crystallography. Molecular Pathology 2000, 53, 8–14, [https://mp.bmj.com/content/53/1/8.full.pdf]. [CrossRef]
- Shi, Y. A Glimpse of Structural Biology through X-Ray Crystallography. Cell 2014, 159, 995–1014. [CrossRef]
- Maveyraud, L.; Mourey, L. Protein X-ray Crystallography and Drug Discovery. Molecules 2020, 25. [CrossRef]
- The Protein Data Bank: Statistics. https://www.rcsb.org/stats. Accessed: 2023-08-15.
- Berman, H.M. et al. The Protein Data Bank. Nucleic Acids Research 2000, 28, 235–242, [https://academic.oup.com/nar/article-pdf/28/1/235/9895144/280235.pdf]. [CrossRef]
- Als-Nielsen, J.; McMorrow, D. Elements of Modern X-ray Physics, 2nd ed.; Wiley: New Jersey, United States, 2011.
- Perutz, M.F. et al. Structure of Hæmoglobin: A Three-Dimensional Fourier Synthesis at 5.5-Å. Resolution, Obtained by X-Ray Analysis. Nature 1960, 185, 416–422. [CrossRef]
- Ramakrishnan, V. et al. Structure of the 30S ribosomal subunit. Nature 2000, 407, 327–339. [CrossRef]
- Garman, E.F. Radiation damage in macromolecular crystallography: what is it and why should we care? Acta Crystallographica Section D 2010, 66, 339–351. [CrossRef]
- Garman, E.F.; Owen, R.L. Cryocooling and radiation damage in macromolecular crystallography. Acta Crystallographica Section D 2006, 62, 32–47. [CrossRef]
- Standfuss, J.; Spence, J. Serial crystallography at synchrotrons and X-ray lasers. IUCrJ 2017, 4, 100–101. [CrossRef]
- Neutze, R. et al. Potential for biomolecular imaging with femtosecond X-ray pulses. Nature 2000, 406, 752–757.
- Chapman, H.N. X-Ray Free-Electron Lasers for the Structure and Dynamics of Macromolecules. Annual Review of Biochemistry 2019, 88, 35–58, PMID: 30601681. [CrossRef]
- Chapman, H. et al. Femtosecond X-ray protein nanocrystallography. Nature 2011, 470, 73–77. [CrossRef]
- Fromme, P.; Spence, J.C. Femtosecond nanocrystallography using X-ray lasers for membrane protein structure determination. Current Opinion in Structural Biology 2011, 21, 509–516. [CrossRef]
- Schlichting, I. Serial femtosecond crystallography: the first five years. IUCrJ 2015, 2, 246–255. [CrossRef]
- Barends, T. R. M. et al. Serial femtosecond crystallography. Nat. Rev. Methods Primers 2022, 2, 59. [CrossRef]
- Wiedorn, M. O. et al. Megahertz serial crystallography. Nat. Communications 2018, 9, 4025. [CrossRef]
- de Wijn, R.; Melo, D.V.M.; Koua, F.H.M.; Mancuso, A.P. Potential of Time-Resolved Serial Femtosecond Crystallography Usin High Repetition Rate XFEL Sources. Appl. Sci. 2022, 12, 2551. [CrossRef]
- Pandey, S.; Poudyal, I.; Malla, T.N. Pump-Probe Time-Resolved Serial Femtosecond Crystallography at X-Ray Free Electron Lasers. Crystals 2020, 10, 628. [CrossRef]
- Aquila, A. et al. Time-resolved protein nanocrystallography using an X-ray free-electron laser. Optics Express 2012, 20, 2706. [CrossRef]
- Kupitz, C. et al. Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser. Nature 2014, 513, 261–265. [CrossRef]
- Orville, A.M. Recent results in time resolved serial femtosecond crystallography at XFELs. Current Opinion in Structural Biology 2020, 65, 193–208. Catalysis and Regulation; Protein Nucleic Acid Interaction. [CrossRef]
- Powell, H.R. X-ray data processing. Bioscience Reports 2017, 37, BSR20170227, [https://portlandpress.com/bioscirep/article-pdf/37/5/BSR20170227/430355/bsr-2017-0227c.pdf]. [CrossRef]
- Kabsch, W. XDS. Acta Cryst. D 2010, 66, 125–132. [CrossRef]
- Battye, T.G.G. et al. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallographica Section D 2011, 67, 271–281. [CrossRef]
- Winter, G. et al. DIALS: implementation and evaluation of a new integration package. Acta Cryst. 2018, D74, 85–97. [CrossRef]
- Brewster, A.S. et al. Improving signal strength in serial crystallography with DIALS geometry refinement. Acta Crystallographica Section D 2018, 74, 877–894. [CrossRef]
- White, T.A. et al. CrystFEL: a software suite for snapshot serial crystallography. J. Appl. Cryst. 2012, 45, 335–341. [CrossRef]
- Lamzin, V.S.; Perrakis, A. Current state of automated crystallographic data analysis. Nature Structural Biology 2000, 7, 978–981. [CrossRef]
- Winter, G.; McAuley, K.E. Automated data collection for macromolecular crystallography. Methods 2011, 55, 81–93. Methods in Structural Proteomics. [CrossRef]
- Perrakis, A.; Morris, R.; Lamzin, V.S. Automated protein model building combined with iterative structure refinement. Nature Structural Biology 1999, 6, 458–463. [CrossRef]
- Alharbi, E. et al. Comparison of automated crystallographic model-building pipelines. Acta Crystallographica Section D Structural Biology 2019, 75, 1119–1128. [CrossRef]
- Hamilton, W.C. The Revolution in Crystallography. Science 1970, 169, 133–141. [CrossRef]
- Abola, E. et al. Automation of X-ray crystallography. Nature Structural Biology 2000, 7, 973–977. [CrossRef]
- Sauter, N.K. XFEL diffraction: developing processing methods to optimize data quality. Journal of Synchrotron Radiation 2015, 22, 239–248. [CrossRef]
- Decking, W. et al. A MHz-repetition-rate hard X-ray free-electron laser driven by a superconducting linear accelerator. Nat. Photonics 2020, 14, 391–397. [CrossRef]
- Tschentscher, Th.. Investigating ultrafast structural dynamics using high repetition rate x-ray FEL radiation at European XFEL. Eur. Phys. J. Plus 2023, 138, 274. [CrossRef]
- Allahgholi, A. et al. The adaptive gain integrating pixel detector. JINST 2016, 11. [CrossRef]
- Hart, M. et al. Development of the LPD, a high dynamic range pixel detector for the European XFEL. 2012 IEEE Nuclear Science Symposium and Medical Imaging Conference Record (NSS/MIC, 2012, pp. 534–537. [CrossRef]
- Mozzanica, A. et al. The JUNGFRAU Detector for Applications at Synchrotron Light Sources and XFELs. Synchr. Rad. News 2018, 31, 16–20. [CrossRef]
- .
- Turkot, O., Dall’Antonia, F. et al. Towards automated analysis of serial crystallography data at the European XFEL. X-Ray Free-Electron Lasers: Advances in Source Development and Instrumentation VI; Tschentscher, T. et al., Ed. International Society for Optics and Photonics, SPIE, 2023, Vol. 12581, p. 125810M. [CrossRef]
- Karplus, P.A.; Diederichs, K. Linking crystallographic model and data quality. Science 2012, 336, 1030–1033. [CrossRef]
- White, T.A. et al. Recent developments in CrystFEL. Journal of Applied Crystallography 2016, 49, 680–689. [CrossRef]
- White, T. CrystFEL: data processing for FEL crystallography. https://www.desy.de/~twhite/crystfel. Accessed: 2023-09-15.
- Dall’Antonia, F., Turkot, O. et al. Code repository of EXtra-Xwiz: pipeline for SFX data analysis at European XFEL. https://github.com/European-XFEL/EXtra-Xwiz/tree/crystals2023. Accessed: 2023-09-17.
- White, T. Processing serial crystallography data with CrystFEL: a step-by-step guide. Acta Cryst. 2019, D75, 1–15. [CrossRef]
- Könnecke, M. et al. The NeXus data format. Journal of Applied Crystallography 2015, 48, 301–305. [CrossRef]
- Bernstein, H.J. et al. Gold Standard for macromolecular crystallography diffraction data. IUCrJ 2020, 7, 784–792. [CrossRef]
- Maia, F.R.N.C. The Coherent X-ray Imaging Data Bank. Nat. Methods 2012, 9, 854–855. [CrossRef]
- Kluyver, T. et al. EXtra-data: library for accessing data at European XFEL. https://extra-data.readthedocs.io. Accessed: 2023-09-15.
- Yoo, A.B.; Jette, M.A.; Grondona, M. SLURM: Simple Linux Utility for Resource Management. Job Scheduling Strategies for Parallel Processing. Springer, 2003, pp. 44–60.
- White, T.A. Post-refinement method for snapshot serial crystallography. Philosophical Transactions of the Royal Society B: Biological Sciences 2014, 369, 20130330. [CrossRef]
- Wrigley, J. et al. DAMNIT: tool for interactive data and metadata inspection at European XFEL. https://rtd.xfel.eu/docs/damnit/en/latest/. Accessed: 2023-05-07.
- Götz, A.; Konrad, U.; Le Gall, E.; Ounsy, M.; Servan, S. VISA sustainability sheet, 2023. [CrossRef]
- Gelisio, L. et al. EuXFEL Data Analysis User Documentation. https://rtd.xfel.eu/docs/data-analysis-user-documentation/en/latest/. Accessed: 2023-09-15.
- Preston-Werner, T. TOML: Tom’s Obvious Minimal Language. https://toml.io/en/. Accessed: 2023-09-15.
- Dall’Antonia, F., Turkot, O. et al. Documentation on EXtra-Xwiz: pipeline for SFX data analysis at European XFEL. https://rtd.xfel.eu/docs/data-analysis-user-documentation/en/latest/software/extra-xwiz/. Accessed: 2023-09-15.
- Kluyver, T. et al. EXtra-geom: library for describing physical layout of multi-module detectors at European XFEL. https://extra-geom.readthedocs.io. Accessed: 2023-05-15.
- White, T. Symmetry Classification for Serial Crystallography Experiments. https://www.desy.de/~twhite/crystfel/twin-calculator.pdf. Accessed: 2023-09-15.
- Le Gall, E. et al. Documentation on VISA: Virtual Infrastructure for Scientific Analysis. https://visa.readthedocs.io/en/latest/index.html. Accessed: 2023-09-15.
- Ferreira de Lima, D.E. et al. Automatic online data analysis optimization: application to serial femtosecond crystallography. International Union of Crystallography 2024. In preparation.
- Yefanov, O. et al. Accurate determination of segmented X-ray detector geometry. Optics Express 2015, 23, 28459. [CrossRef]
| 1 | For an extensive description the reader is referred to, e.g., [6] |
| 2 | For a comprehensive introduction to structure determination using XFELs the reader is referred to [13] |
| 3 | For a detailed explanation, the reader is referred to, e.g., [30] |
| 4 | Hierarchical Data Format v.5 [42] |
| 5 | Detailed description of all available configuration options is available at the EXtra-Xwiz documentation [59] |
Figure 1.
Schematic of EXtra-Xwiz. The input expected by EXtra-Xwiz consists of (i) experimental data, which is represented as a virtual data set (“Data” block), (ii) a detector geometry description (“Geometry” block), and (iii) optionally a unit cell file (“Unit cell” block). Reduction and processing of the data is performed by CrystFEL (“CrystFEL run” block), and input parameters can be either specified in the configuration file or imported from the CrystFEL graphical user interface project file (“CrystFEL GUI proj.” block). The former is executed on the High-Performance Computing (HPC) cluster Maxwell (“HPC cluster” block). Afterwards the output reflections are post-processed by the partialator program from the CrystFEL suite (“partialator run” block), which produces a unique set of structure factors (“SF” file). Reflections can be split by EXtra-Xwiz into custom subsets (“partialator split” block) using conditions derived from the input data (see text for details). Automatic execution of EXtra-Xwiz and harvesting of metadata can be obtained exploiting the DAMNIT tool (“DAMNIT” block). Adapted from [43].
Figure 1.
Schematic of EXtra-Xwiz. The input expected by EXtra-Xwiz consists of (i) experimental data, which is represented as a virtual data set (“Data” block), (ii) a detector geometry description (“Geometry” block), and (iii) optionally a unit cell file (“Unit cell” block). Reduction and processing of the data is performed by CrystFEL (“CrystFEL run” block), and input parameters can be either specified in the configuration file or imported from the CrystFEL graphical user interface project file (“CrystFEL GUI proj.” block). The former is executed on the High-Performance Computing (HPC) cluster Maxwell (“HPC cluster” block). Afterwards the output reflections are post-processed by the partialator program from the CrystFEL suite (“partialator run” block), which produces a unique set of structure factors (“SF” file). Reflections can be split by EXtra-Xwiz into custom subsets (“partialator split” block) using conditions derived from the input data (see text for details). Automatic execution of EXtra-Xwiz and harvesting of metadata can be obtained exploiting the DAMNIT tool (“DAMNIT” block). Adapted from [43].

Figure 2.
Schematic representation of the virtual dataset file (VDS). (a) Data from each module of EuXFEL X-ray detectors is stored as a different file. (b) Data from all modules assembled as a single array in a VDS file. A geometry description ("geom") can then be used to spatially arrange modules in the laboratory frame. In this example, only the eight inner modules are displayed, corresponding to the black file icons in (a).
Figure 2.
Schematic representation of the virtual dataset file (VDS). (a) Data from each module of EuXFEL X-ray detectors is stored as a different file. (b) Data from all modules assembled as a single array in a VDS file. A geometry description ("geom") can then be used to spatially arrange modules in the laboratory frame. In this example, only the eight inner modules are displayed, corresponding to the black file icons in (a).
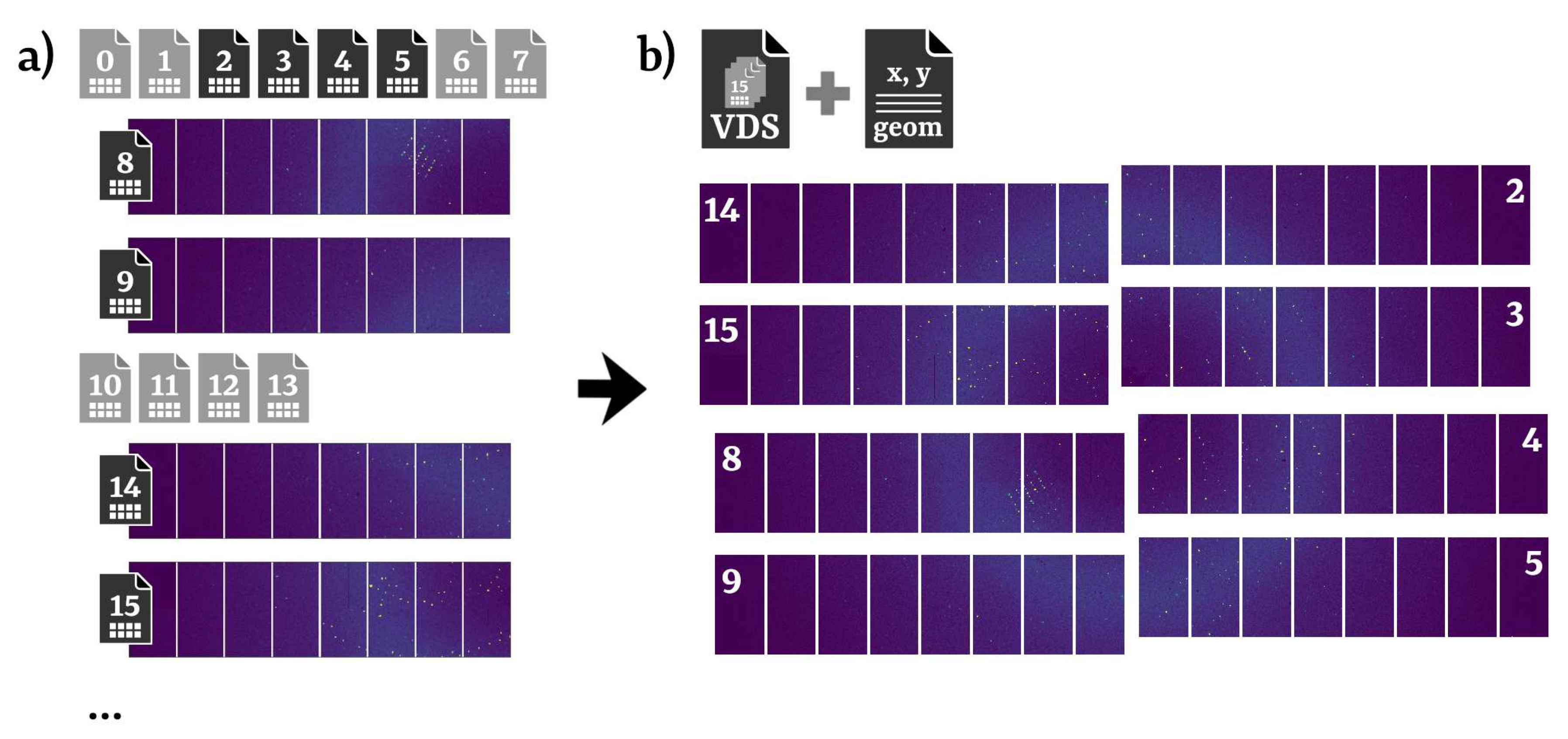
Figure 3.
Schematic of the EXtra-Xwiz workflow used to determine the unit cell in case no initial unit cell file is available. The blocks are the same as defined in Figure 1, except for the cell_explorer graphical user interface displayed in the top right corner.
Figure 3.
Schematic of the EXtra-Xwiz workflow used to determine the unit cell in case no initial unit cell file is available. The blocks are the same as defined in Figure 1, except for the cell_explorer graphical user interface displayed in the top right corner.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Copyright: This open access article is published under a Creative Commons CC BY 4.0 license, which permit the free download, distribution, and reuse, provided that the author and preprint are cited in any reuse.
EXtra-Xwiz: A Tool to Streamline Serial Femtosecond Crystallography Workflows at European XFEL
Oleksii Turkot
et al.
,
2023
Sample Delivery Systems for Serial Femtosecond Crystallography at PAL-XFEL
Jaehyun Park
et al.
,
2023
Achieving a Good Crystal System for Crystallographic X-ray Fragment Screening
Patrick M. Collins
et al.
,
2018
MDPI Initiatives
Important Links
© 2024 MDPI (Basel, Switzerland) unless otherwise stated







