Preprint
Article
Striatal GDNF System Elicits Chemoattraction to RET Positive Dopamine Axons Seven Times Longer in Distance than the Abundant Medium Spiny Neurons and Covers Approximately One Twelfth of the Striatal Neuronal Volume
Altmetrics
Downloads
89
Views
41
Comments
0
A peer-reviewed article of this preprint also exists.
This version is not peer-reviewed
Submitted:
17 May 2024
Posted:
20 May 2024
You are already at the latest version
Alerts
Abstract
Glial cell line-derived neurotrophic factor GDNF is among the strongest dopamine neuron function and survival promoting factors known. Due to this reason, it has clinical relevance in dopamine disorders such as Parkinson’s disease and Schizophrenia. In the striatum, GDNF is exclusively expressed in interneurons which make up only about 0.6% of striatal cells. Despite clinical significance, histological analysis of striatal GDNF system arborization and relevance to incoming dopamine axons, which bear its receptor RET, has remained enigmatic. This is mainly due to the lack of antibodies able to visualize GDNF and RET positive cellular processes; here we overcome this problem by using knock-in marker alleles. We find that GDNF neurons chemoattract RET+ axons at least 7 times longer in distance than medium spiny neurons (MSNs), which make up 95% of striatal neurons. Furthermore, we provide evidence that tyrosine hydroxylase, the rate limiting enzyme in dopamine synthesis, is enriched towards GDNF neurons in the dopamine axons. Finally, we find that GDNF neuron arborizations occupy approximately twelve times less striatal volume than 135 times more abundant MSNs. Collectively, our results improve our understanding of how endogenous GDNF affects striatal dopamine system function.
Keywords:
Subject: Biology and Life Sciences - Neuroscience and Neurology
1. Introduction
GDNF is among, if not the most, potent striatal dopamine function-promoting secreted growth factor known [1,2,3]. In vitro it acts as a chemoattractant [4,5,6] and promotes neuronal arborization in vivo [7,8,9,10]. Due to the above reasons, decades of work have been invested into pre-clinical and 6 clinical trials addressing GDNF delivery as a potential treatment for Parkinson’s disease, where striatal dopamine fibers and subsequently dopamine neurons in the midbrain substantia nigra gradually die. Those trials have unfortunately been largely unsuccessful, with hope remaining that when dosed and delivered properly, GDNF could still stall PD progression [2,11,12,13].
On the other side of PD stand disorders with excess dopamine or dysfunction of dopamine metabolism. About 50% of patients with Schizophrenia have elevated striatal dopamine [14,15,16,17], and dopamine receptor blockers remain as the only available anti-psychotic drugs [17,18,19,20]. Very recently, it was shown that GDNF is upregulated in a subset of SCZ patients, cerebrospinal fluid and in post-mortem striatum. In mice, similar upregulation of endogenous GDNF expression results in a range of SCZ-like features [21], suggesting that excess GDNF may drive the disease in a subset of SCZ patients. Supporting this idea, methamphetamine, a known enhancer of psychosis risk, also transiently upregulates striatal GDNF expression in various animal models [22,23,24]. Thus, excess GDNF may drive, or contribute to Schizophrenia. Further supporting this idea, dopamine enhancing drugs are well known to increase the risk of psychosis [25,26,27]. Therefore, as with many important regulators and drugs, it is likely that the dose and timing for GDNF are of importance in determining the physiological and pathological response. Further illustrating this, ectopic delivery of GDNF leads to a range of side effects in the dopamine function, depending on timing, levels, and site of ectopic expression in the brain [11,28,29,30,31,32].
Despite the above clinical relevance, visualization of the endogenous striatal GDNF system - evaluation of its arborization, effect on incoming dopamine axons which bear its receptors GFRα1 and RET - has remained largely enigmatic. This is because available antibodies for GDNF, like for many growth factors conserved in evolution, are not sufficiently adequate to allow such histochemical analysis. Similarly, antibodies able to stain GDNF receptor RET-positive axons in the striatum are not available [33,34]. Thus, existing knowledge on GDNF expression is based on in situ hybridization or Lac-Z knock-in results into Gdnf locus, which reveal only expressing cell bodies [21,35,36,37] (Figure 1B). Those experiments reveal that only about 0.6% of striatal neurons express GDNF. Those are mainly parvalbumin expressing interneurons with 83% overlap with GDNF, while the remaining GDNF expressing interneurons co-express somatostatin or acetylcholin [36]. If and how GDNF expressing cells spatially influence the incoming dopamine fibers, and how much of striatal arborizations is GDNF positive compared to MSNs has remained unknown. Here, we set to visualize the striatal GDNF system and analyze its interaction with dopamine axons. To overcome the issue of the lack of high-quality antibodies recognizing GDNF and RET, we utilized knock-in alleles Gdnf-CreERT2 [38] in combination with flox-stop-flox-tdTomato (tdTomato) reporter allele [39] and Ret-eGFP knock-in allele [40]. We then used modern confocal and computational tools to evaluate the interaction and to perform volume estimation.
2. Materials and Methods
2.1. Animals
All animal experiments were conducted according to the 3R principles of the European Union Directive 2010/63/EU governing the care and use of experimental animals and following local laws and regulations (Finnish Act on the Protection of Animals Used for Scientific or Educational Purposes (497/2013), Government Decree on the Protection of Animals Used for Scientific or Educational Purposes (564/2013). The protocols were authorized by the national Animal Experiment Board of Finland (license number ESAVI/12046/04.10.07/2017). The knock-in lines Gdnf-CreERT2 and Ret-eGFP and the reporter line flox-stop-flox-tdTomato (tdTomato) were obtained from Jackson Laboratory (RRID:IMSR_JAX:024948, RRID:IMSR_JAX:029847 and RRID:IMSR_JAX:007909 respectively). All mice were maintained in a 129Ola/ICR/C57bl6 mixed genetic background. The mice were group-housed in a specific pathogen-free stage with ad libitum access to food and water under a 12-h light-dark cycle (lights on at 6 a.m.) at a relative humidity of 50-60 % and at a room temperature of 21±1 °C. Bedding (aspen chips, Tapvei) and nest material (Tapvei) were changed weekly, and wooden blocks (Tapvei) were provided for enrichment.
2.2. Tamoxifen Injection
The knock-in alleles Ret-eGFP and Gdnf-CreERT2 were used to identify RET- and GDNF-expressing neurons, respectively (Figure 2A). A female mouse triple-heterozygous for Gdnf-CreERT2, Ret-eGFP and tdTomato was crossed with a male heterozygous for tdTomato. The morning when a vaginal plug was found was designated as embryonic day 0. The pregnant dam was injected i.p. with 25 mg/kg of tamoxifen (TMX) on embryonic days 15 and 16 to induce flox-stop-flox-tdTomato recombination on GDNF-expressing cells. On the day of birth (designated postnatal day 0, P0), the pups were placed with a foster mother until weaned and genotyped on P21. Animals triple-heterozygous for Ret-eGFP, Gdnf-CreERT2 and tdTomato were used for the experiments.
2.3. Isolation of Tissues
Brains were isolated from triple heterozygous mice at 2 months of age. Mice were anesthetized with pentobarbital (Mebunat, 200 mg/kg, i.p., Yliopiston Apteekki) and perfused with warm PBS followed by 4% paraformaldehyde (Sigma). Samples were post-fixed in 4% paraformaldehyde in PBS for 24 hours at room temperature and dehydrated in 30% sucrose (Thermo Fisher Scientific) in PBS prior to sectioning. Sucrose-dehydrated tissues were cryosectioned to a thickness of 40 µm and stored in cryopreservant buffer containing 30% ethylene glycol (Fisher) and 20% glycerol (Acros Organics) in phosphate buffer at -20 °C until analysis.
2.4. Immunohistochemistry
40 µm cryosections containing striatum (STR) or substantia nigra (SN) were permeabilized in PBS with 0.05% Tween20 (Fisher Scientific) and 0.1% Triton-X (Fisher Scientific) (PBSTT), blocked with 2% normal donkey serum (NDS, Abcam), 50mM Glycine (Fisher Scientific) and 0.01% bovine serum albumin (Fisher Scientific) in PBSTT, and stained with primary antibodies diluted in PBSTT with 10mM glycine and 0.1% hydrogen peroxide at +4°C overnight. To visualize tdTomato signal in GDNF expressing cells we used anti-RFP antibodies rabbit anti-RFP (1:500; Rockland CAT: 600-401-379) and goat anti-RFP (1:500; Rockland CAT: 200-101-379). To detect RET expressing cells and fibers we used goat anti-GFP (1:1000; Abcam). Additionally, we used rat anti-DARPP-32 (1:1000, R&D systems CAT MAB4230), rabbit anti-PV (1:1000; Swant CAT PV 27) and rabbit anti-TH (1:1000; Millipore CAT AB152) for DARPP-32, PV and TH detection, respectively. The secondary antibodies used were donkey anti-goat Alexa-Fluor 488, donkey anti-rabbit Alexa-Fluor 488, donkey anti-rabbit Alexa-Fluor-594, donkey anti-rat Alexa-Fluor 488, donkey anti-goat Alexa-Fluor 594 all 1:1000 (Abcam).
2.5. Imaging
Imaging of negative controls and test samples of STR, as well as SN imaging for cell counting were performed using a Zeiss Axio Imager widefield inverted microscope using Zeiss Zen 2 acquisition software. The objective used was a 40x Plan Apochromat 0.8 NA or 20x for the tile images. Samples were imaged with 470/40 nm (38HE GFP), and 587/25nm (64HE mPlum) excitation filters and 525/50nm and 647/70nm emission filters. Images were acquired by tile acquisition using a Hamamatsu ORCA-Flash 4.0 LT B&W camera.
Stained striatal sections for chemoattraction, colocalization and volume analyses were imaged using an inverted confocal microscope (Leica Stellaris 8 FALCON/DLS) using Leica LAS X acquisition software. The objective used was HC PL APO CS2 63x/1.40 with oil immersion. eGFP was imaged with a 499 nm laser and emission window at 504nm-564nm while RFP was imaged with a 590 nm laser with emission window at 595nm-750nm, and detectors used were SiPM (595nm-750nm) and HyD (564nm-570nm). For triple stainings, imaging was done using 499 nm, 579nm and 653 nm lasers, and detectors HyD (504nm-565nm), SiPM (584nm-632nm) and HyD (667nm-750nm). The pinhole was set to 95.5 µm with bidirectional imaging and scan speed 600 Hz. 3D images were acquired by taking a z-stack of approximately 55-60 slices with 0.2 µm spacing with a pixel size of 0.299 µm. Due to the relatively low recombination rate (8%), we imaged every GDNF-expressing neuron from 9 and 6 sections from two different mice, independently of their dorsal-ventral position, until twenty neurons per mouse and a total of 40 neurons were obtained. The imaged neurons spanned all sections from rostral to caudal and therefore the selection of neurons was considered unbiased. The imaging was done in a way that the totality of the neuronal tree visible under the microscope was included into the image. The same approach was used to image PV and DARPP-32 expressing neurons.
2.5. Analysis of Chemoattraction
The analysis was performed using Imaris 10.1. software. To increase the accuracy of our analyses, all images were first processed for background subtraction in each channel using the imaging processing tool in Imaris using a filter of 46.2 um. Briefly, the software smooths the image with an intensity of a Gaussian filtered channel (Gaussian filtered by ¾) minus the intensity of the original channel Gaussian filtered by 8/9 of sphere radius. After background subtraction, the surface of GDNF and DARPP-32 neurons was traced manually using the Imaris “surface” tool (Figure 3Ad,f and Figure 4B and Supplementary Video S2) using a surface detail of 0.145 nm and absolute intensity threshold. The tool “spots” was used to represent Ret-eGFP+ or TH+ fibers on the green channel (Figure 3Ae–f, Supplementary Video S3). Spot detection was set to a diameter of 0.723 µm corresponding to the smallest diameter of the Ret-eGFP or TH axons and using background subtraction as a method for detection. To analyze the spots-surface correlation, we used the spatial view on “Vantage” where the software identified and counted the number of spots close to the neuron surface of interest (Figure 3A g’–g’’’, Supplementary Video S4). To investigate if the distribution of Ret-eGFP+ or TH+ axons was biased towards the surface of GDNF+ or DARPP-32+ neurons (indicating chemoattraction), we analyzed if the number of spots close to the neurons of interest were higher than what would be expected by chance. The tool “spatial random” in the “Vantage” tool on Imaris allowed us to obtain an average of the spots close to neuronal surface from 1000 simulations (dashed line on Figure 3B). The simulations show the distribution of the spots found in each image when the spots would be located randomly (non-biased distribution showing no chemoattraction). We then calculated the percentage of spots obtained from our samples relative to the number of spots obtained from the simulations.
2.6. Analysis of Colocalization of Ret-eGFP and TH in the Striatal Axons
Confocal images of 35 regions of interest (ROI) of size 200 µm x 200 µm were obtained from dorsal to ventral regions of a single striatal section double stained for eGFP and TH. After background subtraction using Imaris (See Section 2.5), the total volume of the green channel (ReteGFP fibers) or red channel (TH fibers) were selected using the “surface” tool. The overlapping volume between the channels was obtained using Imaris Xtension “Surface-Surface Overlap”. The overlapping volume of the red channel over the green channel, and the overlapping volume of the green channel over the red channel were obtained separately for each ROI. We then calculated the percentage of overlapping volumes in the whole striatum (all ROIs), and in the dorsal and ventral striatum separately to investigate whether these regions show a different co-expression pattern of Ret-eGFP and TH.
2.7. Analysis of Colocalization of Ret-eGFPt and TH in the Substantia Nigra
To critically evaluate the results obtained from the Ret-eGFP+ and TH+ fiber volume overlap in the striatum, we stained three sections of the SN from the same knock-in animals used for the rest of the analyses with antibodies against eGFP and TH using the same MM as for the striatal analysis. The number of cells double positive for Ret-eGFP/TH were counted on each hemisphere (two images per section; n=18) from widefield microscopic images using the software ZEISS ZEN 3.3. The overlapping volume of Ret-eGFP and TH fibers was estimated from confocal images using the same methods described in Section 2.6.
2.8. Analysis of Volume of PV and DARPP-32 Neuron Arborization
Confocal images obtained from 5 striatal sections stained for PV and 5 sections stained for DARPP-32 were used for this analysis. A total of 19 images per group were analyzed. Using Imaris we selected ROIs of the same size for all PV and DARPP-32 images (185 µm x 185 µm x 5 µm) to have an accurate estimation of the neuronal volume within the same sampling volume. The total volume of the red channel (PV neurons) or the green channel (DARPP-32 neurons) was selected from each image using the “surface” tool; the surface included both cell bodies and processes. The percentage volume occupied by each type of neuron was then calculated, considering the sum of DARPP-32 and PV neurons as 100%.
2.9. Statistical Analyses
All values are presented as mean ± standard error of the mean (SEM). Statistical comparison between two groups was performed using unpaired Student’s t-test with two-tailed distribution. Multiple comparisons were performed with two-way analysis of variance (ANOVA), followed by Sidak’s post-hoc test. Statistical analysis was performed with GraphPad Prism v10. The statistical significance level was set at p<0.05.
3. Results
3.1. Analysis of Chemoattraction between GDNF Neurons and RET+ Fibers
DA neurons arborize massively in the striatum with axonal tree of one DA neuron covering about 2.7% of the rodent striatum [41] (Figure 1A). DA neurons are also known to express GDNF receptors GFRα1 and RET [42] (Figure 1Ai). GDNF is expressed by striatal interneurons and can be visualized using in situ hybridization [21,37] (Figure 1B) or using LacZ knock-in allele [35,36]. However, antibodies which reliably detect endogenous GDNF and RET and allow analysis of endogenous GDNF system are not available. Thus, we utilized knock-in alleles for Gdnf and Ret as depicted on Figure 2A and immunohistochemical detection of RFP and eGFP (Figure 2B,C and Supplementary Figure S1; see MM for details). We observed RFP expression on scattered neurons within the STR but not in any other brain structure in striatal coronal slices (Figure 2B and Supplementary Figure S1C), while eGFP was observed in cell bodies within the medial septum (MS) and in striatal axons (Figure 2C and Supplementary Figure S1C). We did not observe neither RFP or eGFP expression in any of the negative controls (Supplementary Figure S1A,B). To have a reference of the recombination rate of flox-stop-flox-tdTomato in Gdnf-CreERT2 expressing neurons, we counted the total number of PV+ and GDNF+ neurons in three consecutive STR sections and compared the number of GDNF+ neurons in our samples to the expected number of GDNF+ neurons considering that around 83% of all PV neurons express GDNF [36]. The calculated recombination rate was 8%. On the other hand, around 3% and 4% of the neurons expressing GDNF, also co-express ChAT and Somatostatin (SS), respectively. To gain insight whether flox-stop-flox-tdTomato recombination occurred preferentially in certain GDNF+ neuronal type, we performed triple-stain analysis using antibodies anti-RFP (Gdnf-CreERT2 neurons), anti-GFP (Ret-eGFP) and anti-PV. We found that 95.23% of the analyzed neurons (n=20) were double positive for GDNF and PV, and one neuron (4.76%) was positive only for GDNF. Thus, our results report mainly the biology of PV+ GDNF neurons.
To investigate if GDNF+ neurons attract RET+ fibers, we analyzed RET+ fibers using Ret-eGFP knock-in mice [40] (Figure 3Ab,c). Fibers visible in one microscopic image were defined as “spots” in Imaris software (see MM for details) (Figure 3Ae,f). We analyzed whether the Ret-eGFP+ spots (RET+ fibers) distributed randomly or accumulated near the surface of GDNF+ neurons (Figure 3Ad,f and g’–g’’’). Distribution analysis revealed a peak in the number of eGFP+ spots within about 5 µm from the GDNF neuron surface, after which the number of spots decreased as a function of the distance (Figure 3B,C). To elucidate if the distribution of spots near the neuron surface was higher than what would be expected by chance, we compared the results obtained in each sample (red line in Figure 3B,C) to the average obtained from 1000 simulations which reflect random distribution on the same sample (grey dotted lines in Figure 3B,C) using Imaris software. The average percentage of spots obtained from the analyzed samples is shown in Figure 3C (red line). We found a significant interaction (¤¤¤¤ p<0.0001) between the datasets (test spots and simulated spots) and the distance to GDNF neuron surface. A Sidák’s multiple comparisons test revealed a significant increase in the number of spots at 0-5 µm (around 120%, #### p<0.001). At 5-10 µm, the number of spots decreased, but were on average higher than the average of the simulations. From the distance of 10 µm, the number of spots decreased and was significantly lower than that observed in the simulations. The peak within 5 µm and the reduction of Ret-eGFP+ spots at further distances could indicate that RET+ fibers tend to localize close to the GDNF neuron surface, therefore reducing the number of fibers further away from the neuron. To study in more detail at which distance the Ret-eGFP fibers are chemoattracted towards GDNF neurons, we performed an analysis per µm within 15 µm from the GDNF neurons. We found that that RET+ fibers are significantly chemoattracted to GDNF neurons until 7 µm from GDNF neuronal surface (Figure 3D).
3.2. Analysis of Chemoattraction between RET+ Axons and Medium Spiny Neurons (MSNs)
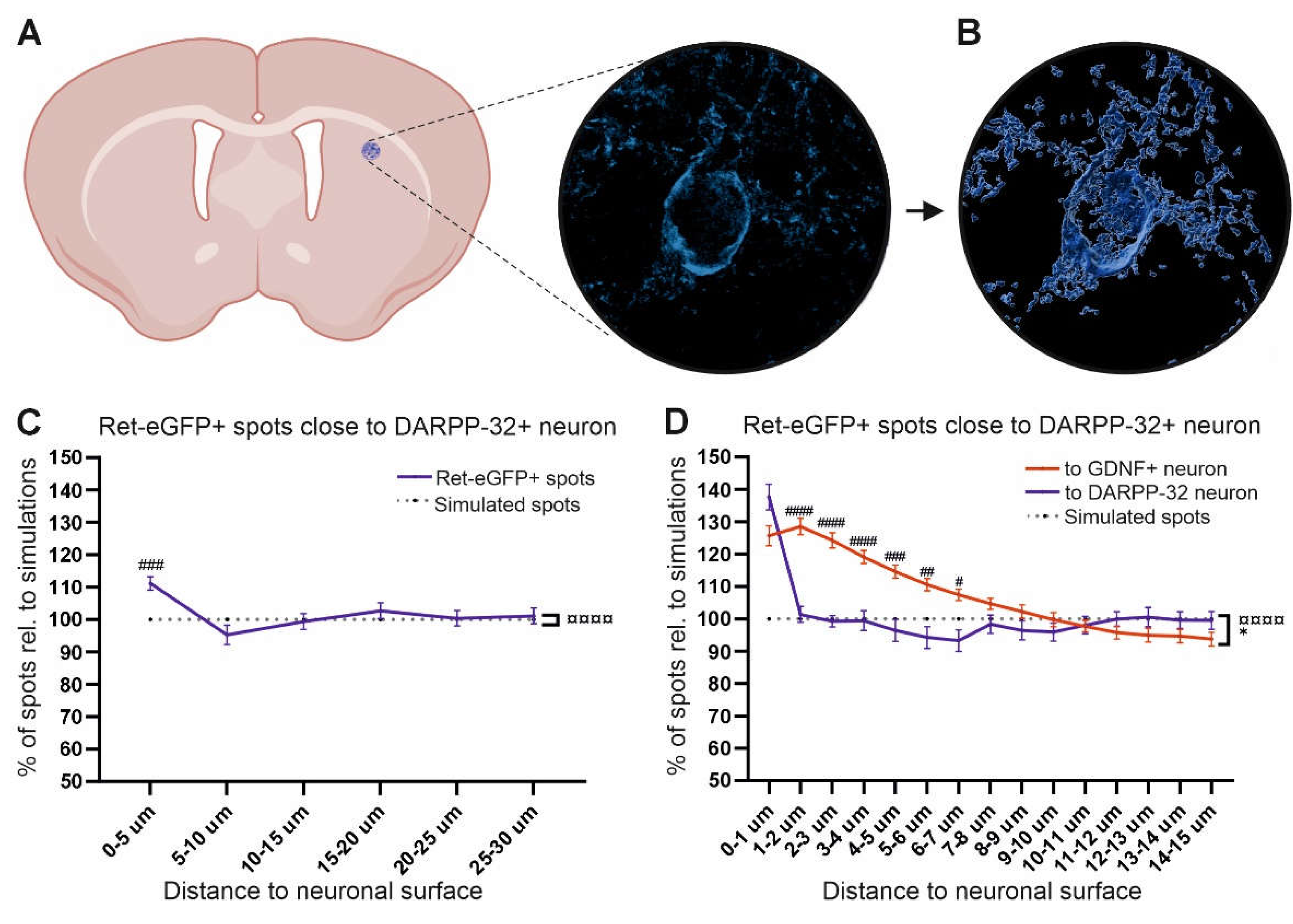
Next, we analyzed the accumulation of RET+ fibers around MSNs, the most abundant striatal neurons which do not express GDNF but are also innervated by dopaminergic axons. MSNs express the marker DARPP-32 which we used for immunohistochemical visualization (Figure 4A). We observed a significant increase in Ret-eGFP positive spots at 0-5 µm of distance (Figure 4C). Analysis of the number of spots per µm from 0-15 µm and comparison to the same data from GDNF neurons revealed that the number of Ret-eGFP+ spots close to MSNs is increased similarly to GDNF neurons at 1 µm distance; however, the number of Ret-eGFP+ spots at 2-7 µm from GDNF+ neurons is significantly higher than the number of Ret-eGFP+ spots at the same distance from DARPP-32 neurons (Figure 4D, compare red line and blue line respectively). With the other words, RET+ axons are chemoattracted within 1 µm from MSNs and within 7 µm from GDNF neurons.
Figure 4.
Analysis of chemoattraction between MSNs (DARPP-32) neurons and RET+ DA neurons. (A) Microscopic 2D view of a DARPP-32 neuron in the STR. (B) 3D view of the surface of the DARPP-32 neuron shown in A. (C) Average percentage of Ret-eGFP+ spots at 0-30 µm from the DARPP-32 neuronal surface. A Two-way ANOVA analysis revealed a significant interaction between independent variables (¤¤¤¤ p<0.0001); main effects showed no difference in the number of spots close to the surface between groups of analysis. However, Sidák´s multiple comparison test showed a significant increase in the number of spots at 0-5 µm (###, p<0.001). (D) Analysis per µm of Ret-eGFP+ spots at 0-15 µm from the DARPP-32 neuron (blue line) compared to Ret-eGFP+ spots close to GDNF+ neurons (red solid line). The number of Ret-eGFP+ spots close to DARPP-32 neurons was similar to GDNF+ neurons at 0-1 µm; however, the number of spots close to GDNF+ neurons was significantly higher at 2-7 µm compared to DARPP-32 neurons. Data is shown as mean ± SEM; significant main effects * p<0.05 and significant interaction between variables ¤¤¤¤ p<0.0001 as obtained by a Two-way ANOVA followed by a Sidák´s multiple comparison test to analyze specific distances (# p<0.05; ## p<0.01; ### p<0.001; #### p<0.0001).
Figure 4.
Analysis of chemoattraction between MSNs (DARPP-32) neurons and RET+ DA neurons. (A) Microscopic 2D view of a DARPP-32 neuron in the STR. (B) 3D view of the surface of the DARPP-32 neuron shown in A. (C) Average percentage of Ret-eGFP+ spots at 0-30 µm from the DARPP-32 neuronal surface. A Two-way ANOVA analysis revealed a significant interaction between independent variables (¤¤¤¤ p<0.0001); main effects showed no difference in the number of spots close to the surface between groups of analysis. However, Sidák´s multiple comparison test showed a significant increase in the number of spots at 0-5 µm (###, p<0.001). (D) Analysis per µm of Ret-eGFP+ spots at 0-15 µm from the DARPP-32 neuron (blue line) compared to Ret-eGFP+ spots close to GDNF+ neurons (red solid line). The number of Ret-eGFP+ spots close to DARPP-32 neurons was similar to GDNF+ neurons at 0-1 µm; however, the number of spots close to GDNF+ neurons was significantly higher at 2-7 µm compared to DARPP-32 neurons. Data is shown as mean ± SEM; significant main effects * p<0.05 and significant interaction between variables ¤¤¤¤ p<0.0001 as obtained by a Two-way ANOVA followed by a Sidák´s multiple comparison test to analyze specific distances (# p<0.05; ## p<0.01; ### p<0.001; #### p<0.0001).
3.3. Analysis of Tyrosine Hydroxylase Distribution in Striatal Ret-eGFP Fibers
In the striatum, the tyrosine kinase receptor RET is known to be expressed exclusively in the axons of midbrain DA neurons [33,34,35,36,37,43]. Tyrosine hydroxylase (TH) is the rate-limiting enzyme in DA synthesis and a commonly used marker to visualize striatal DA axons. However, whether TH is evenly distributed in the striatal DA axons has remained unknown. In Ret-eGFP mice eGFP is expressed in DA neurons and eGFP is commonly known to freely diffuse in the cells [44] thus acting as a dye to visualize DA axons. We analyzed the overlapping volume between Ret-eGFP+ axons and TH in 5 striatal sections (Figure 5A). We found that 55.2% of the total volume covered by Ret-eGFP+ axons overlapped with the TH+ volume (Ret-eGFP+ TH+ on Figure 5B). The remaining 44.8% of the Ret-eGFP+ fiber volume did not express TH. On the other hand, 97.9% of the total TH+ fiber volume co-expresses Ret-eGFP (TH+ Ret-eGFP+ on Figure 5C), and only 2.1% of TH+ fiber volume does not express Ret-eGFP. Thus, while the majority of the dopaminergic axons (TH+) co-express RET, only about half of Ret-eGFP+ striatal axon volume contains TH indicating regulated, not freely diffusing distribution for striatal TH. This spatially specific distribution of TH, compared to the more ubiquitous distribution of eGFP was further analyzed on Figure 5Ad–i. Figure 5Aa,d,g reveal that the lack of TH is restrained to certain areas within the same Ret-eGFP+ fibers, (compare g, h and i on Figure 5A) suggesting that differences in eGFP and TH staining do not stem from differences in antibody diffusion rate or other IHC related artifacts as TH is seen in distinct regions in the same Ret-eGFP+ fiber. An analysis of dorsal versus ventral striatum showed no obvious difference in spatial distribution of TH in Ret-eGFP+ axons between these striatal regions (Figure 5D,E).
Figure 5.
Analysis of Ret-eGFP+ and TH+ fiber volume colocalization. (A) Confocal images of Ret-eGFP+ axons (green), TH+ axons (red) and the merged images. The white arrows show fibers that express both Ret-eGFP and TH in a clearly different pattern. A close-up view (a-c) shows that while Ret-eGFP is more ubiquitously distributed, TH is expressed only in specific sites within the same fibers. The analysis of the volumes selected with Imaris makes this difference more evident in d-i. (B) Volume of Ret-eGFP+ fibers that express TH in the STR. Around 55% of the Ret-eGFP+ axonal volume overlaps with TH+ axonal volume. The rest (44.8%) represents the volume of Ret-eGFP+ axons that are not TH+. (C) Volume of TH+ fibers that overlaps with Ret-eGFP. Around 98% of the TH+ axonal volume overlaps with Ret-eGFP+ volume. The remaining 2% represents the TH+ axons that are not Ret-eGFP+. (D) Analysis of Ret-eGFP+ fiber volume that overlaps with TH+ fibers in the dorsal (58.2%) and ventral (47.7%) striatum. (E) Analysis of TH+ fiber volume that overlaps with Ret-eGFP+ fibers in the dorsal (98.2%) and ventral (97.7%) striatum.
Figure 5.
Analysis of Ret-eGFP+ and TH+ fiber volume colocalization. (A) Confocal images of Ret-eGFP+ axons (green), TH+ axons (red) and the merged images. The white arrows show fibers that express both Ret-eGFP and TH in a clearly different pattern. A close-up view (a-c) shows that while Ret-eGFP is more ubiquitously distributed, TH is expressed only in specific sites within the same fibers. The analysis of the volumes selected with Imaris makes this difference more evident in d-i. (B) Volume of Ret-eGFP+ fibers that express TH in the STR. Around 55% of the Ret-eGFP+ axonal volume overlaps with TH+ axonal volume. The rest (44.8%) represents the volume of Ret-eGFP+ axons that are not TH+. (C) Volume of TH+ fibers that overlaps with Ret-eGFP. Around 98% of the TH+ axonal volume overlaps with Ret-eGFP+ volume. The remaining 2% represents the TH+ axons that are not Ret-eGFP+. (D) Analysis of Ret-eGFP+ fiber volume that overlaps with TH+ fibers in the dorsal (58.2%) and ventral (47.7%) striatum. (E) Analysis of TH+ fiber volume that overlaps with Ret-eGFP+ fibers in the dorsal (98.2%) and ventral (97.7%) striatum.

3.4. Analysis of Colocalization of Ret-eGFP and TH in the Substantia Nigra
To further critically evaluate our methodology, we next analyzed the expression of both TH and RET in the SN of the same mice using antibodies anti-eGFP and anti-TH and the same MM used for analyzing the STR (Figure 6A). We found that 98% of the cells analyzed expressed both Ret-eGFP and TH and only 2% expressed Ret-eGFP but not TH (Figure 6B). Importantly, we did not find a single TH+ cell lacking Ret-eGFP expression. To investigate whether TH localizes within Ret-eGFP neurons in a similar manner as it localizes in the fibers within the STR, we also analyzed the overlapping volumes of Ret-eGFP and TH using the same MM as in the STR. The analysis showed that 44.1% of the Ret-eGFP volume also co-expresses TH (Figure 6Da), while 91% of TH positive volumes express Ret-eGFP (Figure 6Db). These results concur with our findings in the STR, showing a clustered distribution of TH within Ret-eGFP neurons.
3.5. Analysis of Chemoattraction between GDNF Neurons and TH+ Fibers
Next, we analyzed TH+ spots close to the GDNF neuronal surface. The analysis was done in the same striatal sections where Ret-eGFP analysis was performed using the same MM. We found that similarly to the number of Ret-eGFP+ spots around GDNF neurons, TH+ spots mainly accumulate at 0-5 µm distance from the GDNF neuron, while the amount decreases as a function of the distance (Figure 7A). These results concur with the ones obtained from the analysis of Ret-eGFP+ spots and indicate that GDNF neurons chemoattract dopaminergic axons independent of whether they are visualized by TH or by Ret-eGFP expression. However, per µm analysis of the first 15 µm showed that the increase in the number of TH+ spots is significant from 1 to 8 µm (Figure 7B). We then investigated if the chemoattraction effect of GDNF neurons was comparable between Ret-eGFP+ and TH+ fiber volumes. Our analysis revealed that TH clusters are significantly closer to GDNF neurons compared to freely diffusing eGFP in Ret-EGFP fibers (Figure 7C). Specifically, we found that TH+ spots localize closer to GDNF neurons in a significantly higher frequency at distances 1-3 µm compared to Ret-eGFP+ spots (Figure 7D).
3.6. Analysis of GDNF and MSN Neuronal Arborization Volume in the Striatum
MSNs represent around 95% of the rodent striatal neurons, while about 0.7% are Parvalbumin (PV) positive interneurons [45] (Figure 8A). Approximately 83% of PV interneurons express GDNF while 95% GDNF+ cells are PV+ with the remaining GDNF neurons expressing either acetylcholine or somatostatin [36]. The relatively low number of GDNF-expressing neurons prompted us to investigate their arborization in the striatum as extensive arborization could cover a relatively large striatal volume and thus provide cellular frame for influencing striatal dopaminergic function. In our experiments TMX-induced recombination occurred in only about 8% of Gdnf-CreERT2;tdTomato cells (see MM for details). Thus, as the best proxy for GDNF+ system volume we investigated PV+ cellular volume and compared it to the volume of DARPP-32+ signal (Figure 8Ca–d). We found that the striatal volume occupied by DARPP-32+ volume is about 12 times larger compared to PV+ volume (Figure 8B,D).
4. Discussion
Striatal dopamine regulates reward, motivation, movement, attention, and excess dopamine defines psychosis and Schizophrenia (SCZ) probably in at least about 50% of patients [14,15,16,17,46]. Conversely, pro-dopaminergic drugs like L-DOPA increase the risk of psychosis during treatment of Parkinson’s disease (PD) where dopamine (DA) and DA neurons are gradually lost [47]. It is therefore important to understand the mechanisms that regulate dopamine function in the striatum including building the understanding on involved cellular networks architecture and their interactions.
GDNF is a strong dopamine function enhancing factor expressed specifically in only about 0,6% of striatal cells, namely in the striatal interneurons [36,48]. Recent evidence suggests that abnormally high — about 2-fold increase in endogenous GDNF expression — may drive Schizophrenia in a subgroup of patients [21,22]. On the other hand, when striatal DA is lost in PD, 2-3-fold increase in endogenous GDNF expression may carry potential to treat this disorder [2,49,50].
Plenty of evidence suggests that despite being expressed only in about 0.6% of striatal cells, endogenous GDNF and its levels affect brain dopamine (DA) and related functions in various ways. For example, 40% adult-onset reduction in striatal GDNF reduces amphetamine stimulated behavior and increases DA transporter (DAT) activity [51]. About two-fold increase in endogenous GDNF expression on the other hand also increases DAT activity by about 5-fold rendering mice 5-fold more sensitive to 6-OHDA, a toxin pumped into DA neurons by DAT [48]. GDNF heterozygous knock-out (KO) mice with about 50% reduction in GDNF display increase in extracellular DA levels in the striatum likely reflecting compensatory response [52], earlier age-dependent loss in motor function and TH expression at least in some strains of mice [53], anxiety [54], spatial memory defect [55] and altered striatal response to morphine[56]. Thus, despite being expressed in only about 0.6% of striatal cells, relatively small changes in endogenous GDNF levels have a clear impact on striatal DA system function. However, mostly due to the lack of antibodies enabling reliable detection of endogenous GDNF and it’s signaling receptor RET in DA neurons [33,34] the cell biological basis of the above phenomena has remained unexplored.
First, it is important to understand how striatal GDNF system looks to then evaluate its size and effect on the incoming RET+ dopamine axons. Previous studies have shown GDNF to be a chemoattractant in vivo for a diversity of neuronal types in the central and peripheral nervous system of mice, such as neuronal precursor cells in the rostral migratory stream [4], limb motor axons [6] and enteric neural precursors [57]. However, if and how GDNF neurons elicit chemoattraction to RET-expressing striatal dopamine fibers in vivo has not been explored. We found that RET-expressing fibers accumulate around the surface of GDNF neurons up to a distance of 7 µm, providing evidence that GDNF neurons do elicit chemoattraction upon dopaminergic neuron terminals in the mouse striatum.
We also show that GDNF neurons elicit chemoattraction on RET+ dopaminergic axons at least 7 times longer distance than MSNs, the known innervation target of incoming dopamine axons which make up 95% of all striatal neurons [58,59]. Next, we find that although GDNF neurons are about 135-fold less abundant than MSNs, they cover only about 12-fold smaller striatal volume by their arborizations. Because GDNF neurons chemoattract DA axons at about 7-fold longer distance than MSNs, their effective volume compared to MSNs is about 7 times larger so 7 x 7,61% equaling about 53%. A single DA neuron innervates approximately 2.7% of the total striatal volume which is estimated to contain about 75,000 striatal neurons [41]. Since 0.6% of all striatal neurons are GDNF+ interneurons it follows that one DA neuron axon arborizes within a striatal area which contains about 450 GDNF neurons. Since there are about 7600 DA neurons in the mouse SN [60] innervating one striatal side it follows that the striatal space of 2.7% occupied by one DA neuron axonal tree is simultaneously occupied by axonal trees from over 200 other DA neurons. It is now easy to see how a relatively small, as little as 40% or 50% reduction in GDNF levels or about 2-fold increase in endogenous GDNF levels can have a profound effect on striatal DA system function. Furthermore, PNN is known to be rich of proteoglycans known to bind GDNF with high affinity[61].Thus it is feasible to envision how perineuronal network (PNN) known to surround striatal PV interneurons [62] may act as local GDNF reservoir contributing to chemoattraction.
We also find that the distribution of TH within RET expressing axons is not ubiquitous, but rather confined to specific sites within the same fibers. Further, we find that TH, though about 2 times less abundant than likely the whole dopamine axon filling eGFP, is chemoattracted stronger towards GDNF neuron than eGFP+ fibers indicating active positioning within dopamine axon towards the GDNF neuron. Validating our methodology, in the SN we observe that 98% of Ret-eGFP+ neurons express TH and in essence not a single TH+ cell lacks signal from Ret-eGFP under the same staining conditions used for experiments in the STR. Several previous studies have shown evidence of compartmentalization of dopamine into functional striatal sub-domains [63,64,65,66,67]. Our observation that TH shows a specific pattern of enrichment in small areas within single striatal DA fiber with chemoattraction towards GDNF neurons aligns with the idea of functional compartmentalization of dopamine metabolism. Collectively, this data suggests that next to chemoattraction via RET, GDNF neurons may regulate local functional segregation of DA.
In conclusion, our results suggest how low abundance GDNF neurons which make up only about 0.6% of striatal cells can affect striatal DA system function. Those are 7-fold more powerful chemoattraction of RET+ DA axons compared to the abundant MSNs, via extensive arborization, which covers about one twelfth of the striatal volume providing extensive contact surface with dopamine axons, and via regulation of local accumulation of tyrosine hydroxylase in DA axons. Our work thus reveals cellular basis and biological processes which help to understand how relatively small change in endogenous GDNF levels has profound effect of striatal DA system function [48,50,51,52,53,54,55,56] and encourages future drug development to modulate the GDNF system function to address diverse clinical needs ranging from excess dopamine to its deficit.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Supplementary Figure S1: Testing and validation of the immunohistochemical methods used. Video S1: 3D view of a GDNF neuron and eGFP fibers; Video S2: selection of GDNF neuron surface on IMARIS; Video S3: selection of eGFP fibers as spots on IMARIS; Video S4: counting of eGFP spots close to GDNF neuron surface.
Author Contributions
Conceptualization, A.R.M-R. and J.O.A.; methodology, A.R.M-R. and J.O.A.; validation, A.R.M-R. and T.S.; formal analysis, A.R.M-R.; investigation, A.R.M-R. and J.O.A.; resources, J.O.A.; data curation, A.R.M-R.; writing—original draft preparation, A.R.M-R., T.S. and J.O.A.; writing—review and editing, A.R.M-R. and J.O.A.; supervision, J.O.A.; project administration, J.O.A.; funding acquisition, J.O.A. All authors have read and agreed to the published version of the manuscript.
Funding
J.O.A. was supported by the Academy of Finland (grants no. 297727 and 350678), Sigrid Juselius Foundation, ERA-NET NEURON grant nr 352077, Center of Innovative Medicine (CIMED), Hjärnfonden, Swedish Research Council (grants no. 2019-01578 and 2022-01093), Helsinki Institute of Life Science, and by European Research Council (ERC, grant no. 724922).
Institutional Review Board Statement
The protocols were authorized by the national Animal Experiment Board of Finland (license number ESAVI/12046/04.10.07/2017).
Informed Consent Statement
Not applicable.
Data Availability Statement
All data related to this manuscript are available upon request.
Acknowledgments
Imaging was performed at the Biomedicum Imaging Unit, the University of Helsinki, supported by the Helsinki Institute of Life Science (HiLIFE) and Biocenter Finland.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lin, L.F.; Doherty, D.H.; Lile, J.D.; Bektesh, S.; Collins, F. GDNF: a glial cell line-derived neurotrophic factor for midbrain dopaminergic neurons. Science 1993, 260, 1130–1132. [Google Scholar] [CrossRef] [PubMed]
- Barker, R.A.; Bjorklund, A.; Gash, D.M.; Whone, A.; Van Laar, A.; Kordower, J.H.; Bankiewicz, K.; Kieburtz, K.; Saarma, M.; Booms, S.; et al. GDNF and Parkinson’s Disease: Where Next? A Summary from a Recent Workshop. J Parkinsons Dis 2020, 10, 875–891. [Google Scholar] [CrossRef] [PubMed]
- Ibanez, C.F.; Andressoo, J.O. Biology of GDNF and its receptors - Relevance for disorders of the central nervous system. Neurobiol Dis 2017, 97, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Paratcha, G.; Ibanez, C.F.; Ledda, F. GDNF is a chemoattractant factor for neuronal precursor cells in the rostral migratory stream. Mol Cell Neurosci 2006, 31, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.J.; Worley, D.; Sanicola, M.; Dressler, G.R. The RET-glial cell-derived neurotrophic factor (GDNF) pathway stimulates migration and chemoattraction of epithelial cells. J Cell Biol 1998, 142, 1337–1345. [Google Scholar] [CrossRef] [PubMed]
- Dudanova, I.; Gatto, G.; Klein, R. GDNF acts as a chemoattractant to support ephrinA-induced repulsion of limb motor axons. Curr Biol 2010, 20, 2150–2156. [Google Scholar] [CrossRef] [PubMed]
- Irala, D.; Bonafina, A.; Fontanet, P.A.; Alsina, F.C.; Paratcha, G.; Ledda, F. The GDNF-GFRalpha1 complex promotes the development of hippocampal dendritic arbors and spines via NCAM. Development 2016, 143, 4224–4235. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Martinez, J.M.; Perez-Navarro, E.; Gavalda, N.; Alberch, J. Glial cell line-derived neurotrophic factor promotes the arborization of cultured striatal neurons through the p42/p44 mitogen-activated protein kinase pathway. J Neurosci Res 2006, 83, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Bonafina, A.; Trinchero, M.F.; Rios, A.S.; Bekinschtein, P.; Schinder, A.F.; Paratcha, G.; Ledda, F. GDNF and GFRalpha1 Are Required for Proper Integration of Adult-Born Hippocampal Neurons. Cell Rep 2019, 29, 4308–4319. [Google Scholar] [CrossRef]
- Zurn, A.D.; Winkel, L.; Menoud, A.; Djabali, K.; Aebischer, P. Combined effects of GDNF, BDNF, and CNTF on motoneuron differentiation in vitro. J Neurosci Res 1996, 44, 133–141. [Google Scholar] [CrossRef]
- Georgievska, B.; Kirik, D.; Bjorklund, A. Aberrant sprouting and downregulation of tyrosine hydroxylase in lesioned nigrostriatal dopamine neurons induced by long-lasting overexpression of glial cell line derived neurotrophic factor in the striatum by lentiviral gene transfer. Exp Neurol 2002, 177, 461–474, doi:S0014488602980064 [pii].. [Google Scholar]
- Manfredsson, F.P.; Tumer, N.; Erdos, B.; Landa, T.; Broxson, C.S.; Sullivan, L.F.; Rising, A.C.; Foust, K.D.; Zhang, Y.; Muzyczka, N.; et al. Nigrostriatal rAAV-mediated GDNF overexpression induces robust weight loss in a rat model of age-related obesity. Mol Ther 2009, 17, 980–991. [Google Scholar] [CrossRef]
- Meissner, W.G.; Frasier, M.; Gasser, T.; Goetz, C.G.; Lozano, A.; Piccini, P.; Obeso, J.A.; Rascol, O.; Schapira, A.; Voon, V.; et al. Priorities in Parkinson’s disease research. Nat Rev Drug Discov 2011, 10, 377–393. [Google Scholar] [CrossRef]
- Howes, O.D.; Kambeitz, J.; Kim, E.; Stahl, D.; Slifstein, M.; Abi-Dargham, A.; Kapur, S. The nature of dopamine dysfunction in schizophrenia and what this means for treatment. Arch Gen Psychiatry 2012, 69, 776–786. [Google Scholar] [CrossRef]
- Howes, O.; Bose, S.; Turkheimer, F.; Valli, I.; Egerton, A.; Stahl, D.; Valmaggia, L.; Allen, P.; Murray, R.; McGuire, P. Progressive increase in striatal dopamine synthesis capacity as patients develop psychosis: a PET study. Molecular psychiatry 2011, 16, 885–886. [Google Scholar] [CrossRef]
- Howes, O.D.; Egerton, A.; Allan, V.; McGuire, P.; Stokes, P.; Kapur, S. Mechanisms underlying psychosis and antipsychotic treatment response in schizophrenia: insights from PET and SPECT imaging. Current pharmaceutical design 2009, 15, 2550–2559. [Google Scholar] [CrossRef]
- Kesby, J.P.; Eyles, D.W.; McGrath, J.J.; Scott, J.G. Dopamine, psychosis and schizophrenia: the widening gap between basic and clinical neuroscience. Translational psychiatry 2018, 8, 30. [Google Scholar] [CrossRef]
- Stepnicki, P.; Kondej, M.; Kaczor, A.A. Current Concepts and Treatments of Schizophrenia. Molecules 2018, 23. [Google Scholar] [CrossRef]
- Howes, O.D.; McCutcheon, R.; Owen, M.J.; Murray, R.M. The Role of Genes, Stress, and Dopamine in the Development of Schizophrenia. Biological psychiatry 2017, 81, 9–20. [Google Scholar] [CrossRef]
- Howes, O.; McCutcheon, R.; Stone, J. Glutamate and dopamine in schizophrenia: an update for the 21st century. Journal of psychopharmacology (Oxford, England) 2015, 29, 97–115. [Google Scholar] [CrossRef]
- Matlik, K.; Garton, D.R.; Montano-Rodriguez, A.R.; Olfat, S.; Eren, F.; Casserly, L.; Damdimopoulos, A.; Panhelainen, A.; Porokuokka, L.L.; Kopra, J.J.; et al. Elevated endogenous GDNF induces altered dopamine signalling in mice and correlates with clinical severity in schizophrenia. Mol Psychiatry 2022, 27, 3247–3261. [Google Scholar] [CrossRef] [PubMed]
- Casserly, L.; Garton, D.R.; Montano-Rodriguez, A.; Andressoo, J.O. Analysis of Acute and Chronic Methamphetamine Treatment in Mice on Gdnf System Expression Reveals a Potential Mechanism of Schizophrenia Susceptibility. Biomolecules 2023, 13. [Google Scholar] [CrossRef] [PubMed]
- Morrow, B.A.; Roth, R.H.; Redmond, D.E.; Elsworth, J.D. Impact of methamphetamine on dopamine neurons in primates is dependent on age: implications for development of Parkinson’s disease. Neuroscience 2011, 189, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Valian, N.; Ahmadiani, A.; Dargahi, L. Escalating Methamphetamine Regimen Induces Compensatory Mechanisms, Mitochondrial Biogenesis, and GDNF Expression, in Substantia Nigra. J Cell Biochem 2017, 118, 1369–1378. [Google Scholar] [CrossRef] [PubMed]
- McKetin, R.; Lubman, D.I.; Baker, A.L.; Dawe, S.; Ali, R.L. Dose-related psychotic symptoms in chronic methamphetamine users: evidence from a prospective longitudinal study. JAMA Psychiatry 2013, 70, 319–324. [Google Scholar] [CrossRef] [PubMed]
- McKetin, R.; McLaren, J.; Lubman, D.I.; Hides, L. The prevalence of psychotic symptoms among methamphetamine users. Addiction 2006, 101, 1473–1478. [Google Scholar] [CrossRef]
- Chen, C.K.; Lin, S.K.; Sham, P.C.; Ball, D.; Loh, E.W.; Hsiao, C.C.; Chiang, Y.L.; Ree, S.C.; Lee, C.H.; Murray, R.M. Pre-morbid characteristics and co-morbidity of methamphetamine users with and without psychosis. Psychol Med 2003, 33, 1407–1414. [Google Scholar] [CrossRef]
- Georgievska, B.; Kirik, D.; Bjorklund, A. Overexpression of glial cell line-derived neurotrophic factor using a lentiviral vector induces time- and dose-dependent downregulation of tyrosine hydroxylase in the intact nigrostriatal dopamine system. J Neurosci 2004, 24, 6437–6445. [Google Scholar] [CrossRef]
- Rosenblad, C.; Georgievska, B.; Kirik, D. Long-term striatal overexpression of GDNF selectively downregulates tyrosine hydroxylase in the intact nigrostriatal dopamine system. Eur J Neurosci 2003, 17, 260–270. [Google Scholar]
- Hudson, J.; Granholm, A.C.; Gerhardt, G.A.; Henry, M.A.; Hoffman, A.; Biddle, P.; Leela, N.S.; Mackerlova, L.; Lile, J.D.; Collins, F.; et al. Glial cell line-derived neurotrophic factor augments midbrain dopaminergic circuits in vivo. Brain Res Bull 1995, 36, 425–432, 036192309400224O [pii]. [Google Scholar]
- Hebert, M.A.; Gerhardt, G.A. Behavioral and neurochemical effects of intranigral administration of glial cell line-derived neurotrophic factor on aged Fischer 344 rats. J Pharmacol Exp Ther 1997, 282, 760–768. [Google Scholar] [PubMed]
- Hebert, M.A.; Van Horne, C.G.; Hoffer, B.J.; Gerhardt, G.A. Functional effects of GDNF in normal rat striatum: presynaptic studies using in vivo electrochemistry and microdialysis. J Pharmacol Exp Ther 1996, 279, 1181–1190. [Google Scholar] [PubMed]
- Drinkut, A.; Tillack, K.; Meka, D.P.; Schulz, J.B.; Kugler, S.; Kramer, E.R. Ret is essential to mediate GDNF’s neuroprotective and neuroregenerative effect in a Parkinson disease mouse model. Cell Death Dis 2016, 7, e2359. [Google Scholar] [CrossRef] [PubMed]
- Taraviras, S.; Marcos-Gutierrez, C.V.; Durbec, P.; Jani, H.; Grigoriou, M.; Sukumaran, M.; Wang, L.C.; Hynes, M.; Raisman, G.; Pachnis, V. Signalling by the RET receptor tyrosine kinase and its role in the development of the mammalian enteric nervous system. Development 1999, 126, 2785–2797. [Google Scholar] [CrossRef] [PubMed]
- Ortega-de San Luis, C.; Pascual, A. Simultaneous Detection of Both GDNF and GFRalpha1 Expression Patterns in the Mouse Central Nervous System. Front Neuroanat 2016, 10, 73. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo-Figueroa, M.; Bonilla, S.; Gutierrez, F.; Pascual, A.; Lopez-Barneo, J. GDNF Is Predominantly Expressed in the PV+ Neostriatal Interneuronal Ensemble in Normal Mouse and after Injury of the Nigrostriatal Pathway. J Neurosci 2012, 32, 864–872. [Google Scholar] [CrossRef]
- Golden, J.P.; DeMaro, J.A.; Osborne, P.A.; Milbrandt, J.; Johnson, E.M., Jr. Expression of neurturin, GDNF, and GDNF family-receptor mRNA in the developing and mature mouse. Exp Neurol 1999, 158, 504–528. [Google Scholar] [CrossRef] [PubMed]
- Cebrian, C.; Asai, N.; D’Agati, V.; Costantini, F. The number of fetal nephron progenitor cells limits ureteric branching and adult nephron endowment. Cell Rep 2014, 7, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Madisen, L.; Zwingman, T.A.; Sunkin, S.M.; Oh, S.W.; Zariwala, H.A.; Gu, H.; Ng, L.L.; Palmiter, R.D.; Hawrylycz, M.J.; Jones, A.R.; et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 2010, 13, 133–140. [Google Scholar] [CrossRef]
- Jain, S.; Golden, J.P.; Wozniak, D.; Pehek, E.; Johnson, E.M., Jr.; Milbrandt, J. RET is dispensable for maintenance of midbrain dopaminergic neurons in adult mice. J Neurosci 2006, 26, 11230–11238. [Google Scholar] [CrossRef]
- Matsuda, W.; Furuta, T.; Nakamura, K.C.; Hioki, H.; Fujiyama, F.; Arai, R.; Kaneko, T. Single nigrostriatal dopaminergic neurons form widely spread and highly dense axonal arborizations in the neostriatum. J Neurosci 2009, 29, 444–453. [Google Scholar] [CrossRef]
- Airaksinen, M.S.; Saarma, M. The GDNF family: signalling, biological functions and therapeutic value. Nat Rev Neurosci 2002, 3, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Trupp, M.; Belluardo, N.; Funakoshi, H.; Ibanez, C.F. Complementary and overlapping expression of glial cell line-derived neurotrophic factor (GDNF), c-ret proto-oncogene, and GDNF receptor-alpha indicates multiple mechanisms of trophic actions in the adult rat CNS. J Neurosci 1997, 17, 3554–3567. [Google Scholar] [CrossRef] [PubMed]
- Malinow, R.; Hayashi, Y.; Maletic-Savatic, M.; Zaman, S.H.; Poncer, J.C.; Shi, S.H.; Esteban, J.A.; Osten, P.; Seidenman, K. Introduction of green fluorescent protein (GFP) into hippocampal neurons through viral infection. Cold Spring Harb Protoc 2010, 2010, pdb–prot5406. [Google Scholar] [CrossRef] [PubMed]
- Tepper, J.M.; Tecuapetla, F.; Koos, T.; Ibanez-Sandoval, O. Heterogeneity and diversity of striatal GABAergic interneurons. Front Neuroanat 2010, 4, 150. [Google Scholar] [CrossRef] [PubMed]
- Jauhar, S.; Veronese, M.; Nour, M.M.; Rogdaki, M.; Hathway, P.; Turkheimer, F.E.; Stone, J.; Egerton, A.; McGuire, P.; Kapur, S.; et al. Determinants of treatment response in first-episode psychosis: an (18)F-DOPA PET study. Mol Psychiatry 2019, 24, 1502–1512. [Google Scholar] [CrossRef] [PubMed]
- Samudra, N.; Patel, N.; Womack, K.B.; Khemani, P.; Chitnis, S. Psychosis in Parkinson Disease: A Review of Etiology, Phenomenology, and Management. Drugs Aging 2016, 33, 855–863. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Kopra, J.; Varendi, K.; Porokuokka, L.L.; Panhelainen, A.; Kuure, S.; Marshall, P.; Karalija, N.; Harma, M.A.; Vilenius, C.; et al. GDNF Overexpression from the Native Locus Reveals its Role in the Nigrostriatal Dopaminergic System Function. PLoS Genet 2015, 11, e1005710. [Google Scholar] [CrossRef] [PubMed]
- Ibanez, C.F.; Andressoo, J.O. Biology of GDNF and its receptors - Relevance for disorders of the central nervous system. Neurobiol Dis 2016. [Google Scholar] [CrossRef]
- Espinoza, S.; Scarpato, M.; Damiani, D.; Manago, F.; Mereu, M.; Contestabile, A.; Peruzzo, O.; Carninci, P.; Santoro, C.; Papaleo, F.; et al. SINEUP Non-coding RNA Targeting GDNF Rescues Motor Deficits and Neurodegeneration in a Mouse Model of Parkinson’s Disease. Mol Ther 2020, 28, 642–652. [Google Scholar] [CrossRef]
- Kopra, J.J.; Panhelainen, A.; Af Bjerken, S.; Porokuokka, L.L.; Varendi, K.; Olfat, S.; Montonen, H.; Piepponen, T.P.; Saarma, M.; Andressoo, J.O. Dampened Amphetamine-Stimulated Behavior and Altered Dopamine Transporter Function in the Absence of Brain GDNF. J Neurosci 2017, 37, 1581–1590. [Google Scholar] [CrossRef]
- Airavaara, M.; Planken, A.; Gaddnas, H.; Piepponen, T.P.; Saarma, M.; Ahtee, L. Increased extracellular dopamine concentrations and FosB/DeltaFosB expression in striatal brain areas of heterozygous GDNF knockout mice. Eur J Neurosci 2004, 20, 2336–2344. [Google Scholar] [CrossRef] [PubMed]
- Boger, H.A.; Middaugh, L.D.; Huang, P.; Zaman, V.; Smith, A.C.; Hoffer, B.J.; Tomac, A.C.; Granholm, A.C. A partial GDNF depletion leads to earlier age-related deterioration of motor function and tyrosine hydroxylase expression in the substantia nigra. Exp Neurol 2006, 202, 336–347. [Google Scholar] [CrossRef] [PubMed]
- Glerup, S.; Lume, M.; Olsen, D.; Nyengaard, J.R.; Vaegter, C.B.; Gustafsen, C.; Christensen, E.I.; Kjolby, M.; Hay-Schmidt, A.; Bender, D.; et al. SorLA controls neurotrophic activity by sorting of GDNF and its receptors GFRalpha1 and RET. Cell Rep 2013, 3, 186–199. [Google Scholar] [CrossRef] [PubMed]
- Gerlai, R.; McNamara, A.; Choi-Lundberg, D.L.; Armanini, M.; Ross, J.; Powell-Braxton, L.; Phillips, H.S. Impaired water maze learning performance without altered dopaminergic function in mice heterozygous for the GDNF mutation. Eur J Neurosci 2001, 14, 1153–1163. [Google Scholar]
- Airavaara, M.; Mijatovic, J.; Vihavainen, T.; Piepponen, T.P.; Saarma, M.; Ahtee, L. In heterozygous GDNF knockout mice the response of striatal dopaminergic system to acute morphine is altered. Synapse 2006, 59, 321–329. [Google Scholar] [CrossRef]
- Pawolski, V.; Schmidt, M.H.H. Neuron-Glia Interaction in the Developing and Adult Enteric Nervous System. Cells 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Freund, T.F.; Powell, J.F.; Smith, A.D. Tyrosine hydroxylase-immunoreactive boutons in synaptic contact with identified striatonigral neurons, with particular reference to dendritic spines. Neuroscience 1984, 13, 1189–1215. [Google Scholar] [CrossRef]
- Surmeier, D.J.; Ding, J.; Day, M.; Wang, Z.; Shen, W. D1 and D2 dopamine-receptor modulation of striatal glutamatergic signaling in striatal medium spiny neurons. Trends Neurosci 2007, 30, 228–235. [Google Scholar] [CrossRef]
- Ip, C.W.; Cheong, D.; Volkmann, J. Stereological Estimation of Dopaminergic Neuron Number in the Mouse Substantia Nigra Using the Optical Fractionator and Standard Microscopy Equipment. Journal of visualized experiments : JoVE. [CrossRef]
- Bespalov, M.M.; Sidorova, Y.A.; Tumova, S.; Ahonen-Bishopp, A.; Magalhaes, A.C.; Kulesskiy, E.; Paveliev, M.; Rivera, C.; Rauvala, H.; Saarma, M. Heparan sulfate proteoglycan syndecan-3 is a novel receptor for GDNF, neurturin, and artemin. J Cell Biol 2011, 192, 153–169. [Google Scholar] [CrossRef]
- Wingert, J.C.; Sorg, B.A. Impact of Perineuronal Nets on Electrophysiology of Parvalbumin Interneurons, Principal Neurons, and Brain Oscillations: A Review. Front Synaptic Neurosci 2021, 13, 673210. [Google Scholar] [CrossRef]
- Meszaros, J.; Cheung, T.; Erler, M.M.; Kang, U.J.; Sames, D.; Kellendonk, C.; Sulzer, D. Evoked transients of pH-sensitive fluorescent false neurotransmitter reveal dopamine hot spots in the globus pallidus. Elife 2018, 7. [Google Scholar] [CrossRef]
- Pereira, D.B.; Schmitz, Y.; Meszaros, J.; Merchant, P.; Hu, G.; Li, S.; Henke, A.; Lizardi-Ortiz, J.E.; Karpowicz, R.J., Jr.; Morgenstern, T.J.; et al. Fluorescent false neurotransmitter reveals functionally silent dopamine vesicle clusters in the striatum. Nat Neurosci 2016, 19, 578–586. [Google Scholar] [CrossRef]
- Cartier, E.A.; Parra, L.A.; Baust, T.B.; Quiroz, M.; Salazar, G.; Faundez, V.; Egana, L.; Torres, G.E. A biochemical and functional protein complex involving dopamine synthesis and transport into synaptic vesicles. J Biol Chem 2010, 285, 1957–1966. [Google Scholar] [CrossRef]
- Sulzer, D.; Cragg, S.J.; Rice, M.E. Striatal dopamine neurotransmission: regulation of release and uptake. Basal Ganglia 2016, 6, 123–148. [Google Scholar] [CrossRef]
- Liu, C.; Kershberg, L.; Wang, J.; Schneeberger, S.; Kaeser, P.S. Dopamine Secretion Is Mediated by Sparse Active Zone-like Release Sites. Cell 2018, 172, 706–718. [Google Scholar] [CrossRef]
Figure 1.
Schematic overview of dopamine (DA) and GDNF systems in the striatum (STR). (A) DA neuron projecting from the Substantia nigra pars compacta (SNc). (Ai) Close up scheme of GDNF receptors, RET and GFRα1, located on DA neuron axons. (B) GDNF is expressed in about 0.6% of all striatal neurons. The photo shows Gdnf mRNA expressing cells (arrows) in a coronal section of mouse striatum, detected by in situ hybridization (RNAscope). Note the regular spacing but the low number of Gdnf+ cells.
Figure 1.
Schematic overview of dopamine (DA) and GDNF systems in the striatum (STR). (A) DA neuron projecting from the Substantia nigra pars compacta (SNc). (Ai) Close up scheme of GDNF receptors, RET and GFRα1, located on DA neuron axons. (B) GDNF is expressed in about 0.6% of all striatal neurons. The photo shows Gdnf mRNA expressing cells (arrows) in a coronal section of mouse striatum, detected by in situ hybridization (RNAscope). Note the regular spacing but the low number of Gdnf+ cells.
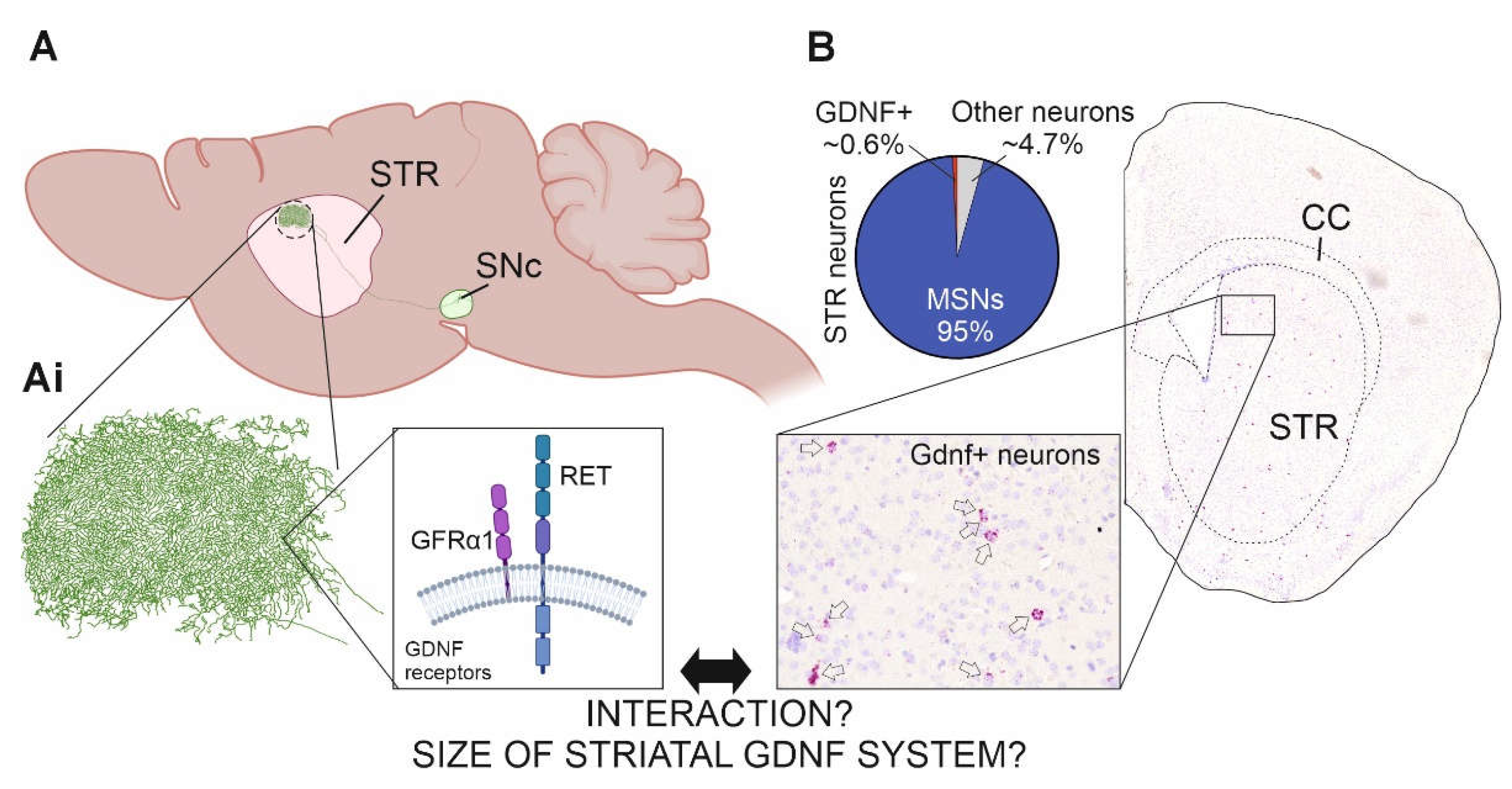
Figure 2.
Experimental setup and timeline. (A) Utilized alleles for the analysis of RET+ axons and GDNF+ neurons in the mouse striatum. The knock-in alleles Ret-eGFP and Gdnf-CreERT2 were used to identify RET- and GDNF-expressing neurons. Tamoxifen was used to induce flox-stop-flox-tdTomato recombination in GDNF expressing cells during days 15 and 16 of gestation; animals triple-heterozygous for Ret-eGFP, Gdnf-CreERT2 and tdTomato were used for the experiments. Ret-eGFP expressing neurons were observed after staining with α-GFP antibody; GDNF expressing neurons were observed after recombination with TMX and staining with α-RFP antibody. (B) Right: microscopic image (widefield) showing RFP positive striatal interneurons in Gdnf-CreERT2 x tdTomato mice; left: confocal image showing a single GDNF expressing neuron. (C) Left: microscopic image (widefield) showing expression of Ret-eGFP in the mouse striatum (STR) and medial septum (MS); right: confocal image eGFP positive axons in the striatum of Ret-eGFP mice is shown on the right.
Figure 2.
Experimental setup and timeline. (A) Utilized alleles for the analysis of RET+ axons and GDNF+ neurons in the mouse striatum. The knock-in alleles Ret-eGFP and Gdnf-CreERT2 were used to identify RET- and GDNF-expressing neurons. Tamoxifen was used to induce flox-stop-flox-tdTomato recombination in GDNF expressing cells during days 15 and 16 of gestation; animals triple-heterozygous for Ret-eGFP, Gdnf-CreERT2 and tdTomato were used for the experiments. Ret-eGFP expressing neurons were observed after staining with α-GFP antibody; GDNF expressing neurons were observed after recombination with TMX and staining with α-RFP antibody. (B) Right: microscopic image (widefield) showing RFP positive striatal interneurons in Gdnf-CreERT2 x tdTomato mice; left: confocal image showing a single GDNF expressing neuron. (C) Left: microscopic image (widefield) showing expression of Ret-eGFP in the mouse striatum (STR) and medial septum (MS); right: confocal image eGFP positive axons in the striatum of Ret-eGFP mice is shown on the right.

Figure 3.
Analysis of chemoattraction between GDNF+ and RET+ DA neurons in the STR. (Aa) RFP in a single Gdnf-CreERT2+ neuron (termed GDNF), (Ab) eGFP in Ret-eGFP+ axons (termed Ret-eGFP), (Ac) merged image. (Ad) GDNF neuron surface traced with Imaris, (Ae) Ret-eGFP fibers represented as spots (Ret-eGFP+ spots). (Ag’,g’’,g’’’) Visualization of eGFP+ spots located at 0-1, 2-3 and 4-5 µm from the GDNF neuron surface, respectively (B) Representative graph of the analysis of the number of Ret-eGFP+ spots close to the surface of a single GDNF neuron, red line shows obtained measurement; grey lines show the expected distribution when Ret-eGFP spots would be randomly distributed (no chemoattraction, see MM for details). (C) Average percentage of Ret-eGFP+ spots close to GDNF neuronal surface at distances 0-30 µm compiled from all samples analyzed (n=40 GDNF neurons). An overall significant interaction between groups and distance was found (¤¤¤¤ p<0.0001). The number of Ret-eGFP+ spots at 0-5 µm from the GDNF+ neurons was significantly higher than expected by chance (#### p<0.0001), and significantly less abundant further away from the neuronal surface from 15-20 µm onwards. (D) Analysis per µm of Ret-eGFP+ spots at 0-15 µm from the GDNF+ neuronal surface revealed a significant difference between groups (**** p<0.0001), and a significant interaction between variables (¤¤¤¤ p<0,0001). A significant increase in Ret-eGFP+ spots was found at distances 1-7 µm from the GDNF neurons. Data is shown as mean ± SEM; significant main effects **** p<0.0001 and significant interaction between variables ¤¤¤¤ p<0.0001 as obtained by a Two-way ANOVA followed by a Sidák’s multiple comparison test to analyze specific distances (### p<0.001; #### p<0.0001).
Figure 3.
Analysis of chemoattraction between GDNF+ and RET+ DA neurons in the STR. (Aa) RFP in a single Gdnf-CreERT2+ neuron (termed GDNF), (Ab) eGFP in Ret-eGFP+ axons (termed Ret-eGFP), (Ac) merged image. (Ad) GDNF neuron surface traced with Imaris, (Ae) Ret-eGFP fibers represented as spots (Ret-eGFP+ spots). (Ag’,g’’,g’’’) Visualization of eGFP+ spots located at 0-1, 2-3 and 4-5 µm from the GDNF neuron surface, respectively (B) Representative graph of the analysis of the number of Ret-eGFP+ spots close to the surface of a single GDNF neuron, red line shows obtained measurement; grey lines show the expected distribution when Ret-eGFP spots would be randomly distributed (no chemoattraction, see MM for details). (C) Average percentage of Ret-eGFP+ spots close to GDNF neuronal surface at distances 0-30 µm compiled from all samples analyzed (n=40 GDNF neurons). An overall significant interaction between groups and distance was found (¤¤¤¤ p<0.0001). The number of Ret-eGFP+ spots at 0-5 µm from the GDNF+ neurons was significantly higher than expected by chance (#### p<0.0001), and significantly less abundant further away from the neuronal surface from 15-20 µm onwards. (D) Analysis per µm of Ret-eGFP+ spots at 0-15 µm from the GDNF+ neuronal surface revealed a significant difference between groups (**** p<0.0001), and a significant interaction between variables (¤¤¤¤ p<0,0001). A significant increase in Ret-eGFP+ spots was found at distances 1-7 µm from the GDNF neurons. Data is shown as mean ± SEM; significant main effects **** p<0.0001 and significant interaction between variables ¤¤¤¤ p<0.0001 as obtained by a Two-way ANOVA followed by a Sidák’s multiple comparison test to analyze specific distances (### p<0.001; #### p<0.0001).

Figure 6.
Analysis of colocalization of Ret-eGFP and TH in the substantia nigra. (A) Widefield images of the SN of the same animals analyzed in Figure 5 showing expression of Ret-eGFP (green), TH (red) and the merged image. (B) Around 98% of the cells analyzed in the SN co-express Ret-eGFP and TH; only 2% express eGFP and not TH, while 0% of cells expressed TH without Ret-eGFP. (C) Confocal images of SN neurons showing expression of Ret-eGFP (green) and TH (red) and the merged images at the top, and their respective volumes selected with Imaris at the bottom. The volumes were used to estimate overlapping volume of each channel. (D) Overlapping volumes of Ret-eGFP and TH: a) Around 44% of the volume expressing Ret-eGFP also coexpressed TH, while around 56% did not express TH; b) around 91% of TH positive volume co-expressed Ret-eGFP, while around 9% did not express Ret-eGFP.
Figure 6.
Analysis of colocalization of Ret-eGFP and TH in the substantia nigra. (A) Widefield images of the SN of the same animals analyzed in Figure 5 showing expression of Ret-eGFP (green), TH (red) and the merged image. (B) Around 98% of the cells analyzed in the SN co-express Ret-eGFP and TH; only 2% express eGFP and not TH, while 0% of cells expressed TH without Ret-eGFP. (C) Confocal images of SN neurons showing expression of Ret-eGFP (green) and TH (red) and the merged images at the top, and their respective volumes selected with Imaris at the bottom. The volumes were used to estimate overlapping volume of each channel. (D) Overlapping volumes of Ret-eGFP and TH: a) Around 44% of the volume expressing Ret-eGFP also coexpressed TH, while around 56% did not express TH; b) around 91% of TH positive volume co-expressed Ret-eGFP, while around 9% did not express Ret-eGFP.

Figure 7.
Analysis of chemoattraction between GDNF neurons and TH+ DA axonal arborizations in the striatum. (A) Average percentage of TH+ spots at distances 0-30 µm from the GDNF neuronal surface. A significant increase on TH+ spots was identified at 0-5 µm (## p<0.01) and to lesser extent at 5-10 µm (# p<0.05) from the neuronal surface. A significant interaction between groups and distance to neuronal surface was found (¤¤¤¤ p<0.0001). (B) Analysis per µm of TH+ spots at 0-15 µm of the neuronal surface. An increase on TH+ spots was found at 0-8 µm. A significant difference between the analyzed groups (*** p<0.001), as well as a significant interaction (¤¤¤¤ p<0.0001) were found. (C) Comparison of the chemoattraction effect of GDNF neurons on Ret-eGFP+ fiber volumes (red line) and TH+ fiber volumes (black line). The data show that TH spots are enriched towards the GDNF neurons. (D) Close up analysis of C (dotted square), showing a significant difference (* p<0.05) between the number of TH+ spots and Ret-eGFP+ spots within the first 3 µm from the GDNF neurons. Data is shown as mean ± SEM; significant main effects * p<0.05, *** p<0.001 and significant interaction between variables ¤¤¤¤ p<0.0001 as obtained by a Two-way ANOVA followed by a Sidák´s multiple comparison test to analyze specific distances (# p<0.05; ## p<0.01; ### p<0.001).
Figure 7.
Analysis of chemoattraction between GDNF neurons and TH+ DA axonal arborizations in the striatum. (A) Average percentage of TH+ spots at distances 0-30 µm from the GDNF neuronal surface. A significant increase on TH+ spots was identified at 0-5 µm (## p<0.01) and to lesser extent at 5-10 µm (# p<0.05) from the neuronal surface. A significant interaction between groups and distance to neuronal surface was found (¤¤¤¤ p<0.0001). (B) Analysis per µm of TH+ spots at 0-15 µm of the neuronal surface. An increase on TH+ spots was found at 0-8 µm. A significant difference between the analyzed groups (*** p<0.001), as well as a significant interaction (¤¤¤¤ p<0.0001) were found. (C) Comparison of the chemoattraction effect of GDNF neurons on Ret-eGFP+ fiber volumes (red line) and TH+ fiber volumes (black line). The data show that TH spots are enriched towards the GDNF neurons. (D) Close up analysis of C (dotted square), showing a significant difference (* p<0.05) between the number of TH+ spots and Ret-eGFP+ spots within the first 3 µm from the GDNF neurons. Data is shown as mean ± SEM; significant main effects * p<0.05, *** p<0.001 and significant interaction between variables ¤¤¤¤ p<0.0001 as obtained by a Two-way ANOVA followed by a Sidák´s multiple comparison test to analyze specific distances (# p<0.05; ## p<0.01; ### p<0.001).

Figure 8.
Analysis of parvalbumin (PV) and DARPP-32 neuron arborization volume in the STR. (A) MSNs represent around 95% of the rodent striatal neurons. Approximately 0.7% of all STR neurons are PV+. (B) Volume covered by PV+ (red) and DARPP-32+ (blue) neurons in ROIs of similar thickness (5 µm). The volume of DARPP-32 was about 12 times higher than that of PV neurons. Data is shown as mean ± SEM, **** p<0.0001 as obtained by a T-student test. (C) Representative confocal images of (a) PV neurons, (c) DARPP-32 neurons, and their analyzed surfaces (b) and (c), respectively. (D) Percentage of the analyzed volume covered by MSNs (92.39%) and PV neurons (7.61%) calculated from B, considering the sum of volumes of PV and DARPP-32 as 100%.
Figure 8.
Analysis of parvalbumin (PV) and DARPP-32 neuron arborization volume in the STR. (A) MSNs represent around 95% of the rodent striatal neurons. Approximately 0.7% of all STR neurons are PV+. (B) Volume covered by PV+ (red) and DARPP-32+ (blue) neurons in ROIs of similar thickness (5 µm). The volume of DARPP-32 was about 12 times higher than that of PV neurons. Data is shown as mean ± SEM, **** p<0.0001 as obtained by a T-student test. (C) Representative confocal images of (a) PV neurons, (c) DARPP-32 neurons, and their analyzed surfaces (b) and (c), respectively. (D) Percentage of the analyzed volume covered by MSNs (92.39%) and PV neurons (7.61%) calculated from B, considering the sum of volumes of PV and DARPP-32 as 100%.

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Copyright: This open access article is published under a Creative Commons CC BY 4.0 license, which permit the free download, distribution, and reuse, provided that the author and preprint are cited in any reuse.
Differences between Cerebral Organoids Derived from a non-PD and a Parkinson’s Patient: A Potential Vulnerability for Neuroinvasive Viruses
Emily Schultz
et al.
,
2021
MDPI Initiatives
Important Links
© 2024 MDPI (Basel, Switzerland) unless otherwise stated








