PCR Methods and applications
- 1. PCR Methods And Thermostable DNA Polymerases Behzad Milani PhD Student of Biochemistry Supervised by Prof. AmirMozaffari November 2016
- 4. Contents of part I: • History of PCR • Polymerase Chain Reaction • Steps involved • Applications of PCR • Factors for optimal PCR • Variations of PCR methods and their applications • Comparison PCR & Cloning • Advantages • Limitations
- 5. History of PCR • In 1983 Kary Mullis, a scientist working for the Cetus Corporation was driving along US Route 101 in northern California when he came up with the idea for the polymerase chain reaction. • In 1985 the polymerase chain reaction was introduced to the scientific community at a conference in October. Cetus rewarded Kary Mullis with a $10,000 bonus for his invention. • Later, during a corporate reorganization, Cetus sold the patent for the PCR process to a pharmaceutical company Hoffmann-LaRoche for $300 million. • In 1993 Mullis awarded nobel prize in Chemistry along with Michael Smith for his work on PCR.
- 6. Polymerase Chain Reaction • PCR targets and amplifies a specific region of a DNA strand. • It is an invitro technique to generate large quantities of a specified DNA. • Often, only a small amount of DNA is available eg.A drop of blood, Semen strains, Single hair, vaginal swabs etc. • Two methods currently exist for amplifying the DNA or making copies Cloning—takes a long time for enough clones to reach maturity PCR—works on even a single molecule quickly
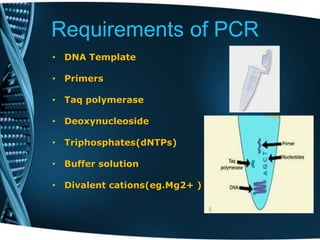
- 8. Requirements of PCR • DNA Template • Primers • Taq polymerase • Deoxynucleoside • Triphosphates(dNTPs) • Buffer solution • Divalent cations(eg.Mg2+ )
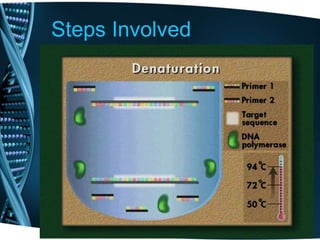
- 10. Steps Involved Denaturation • The reaction mixture is heated to a temperature between 90-98°C so that the ds DNA is denatured into single strands by disrupting the hydrogen bonds between complementary bases. • Duration of this step is 1-2 mins.
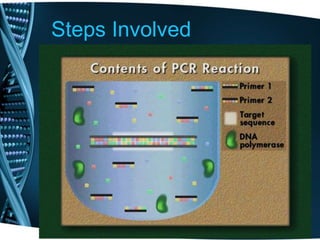
- 11. Steps Involved
- 12. Steps Involved Annealing • Temperature of reaction mixture is cooled to 45-60°C • Primers are jiggling around caused by ??????? • Primers base pair with the complementary sequence in the DNA. • Hydrogen bonds reform. • Annealing fancy word for renaturing.
- 13. Steps Involved
- 14. Steps Involved Extension • The temperature is now shifted to 72°C which is ideal for polymerase. • Primers are extended by joining the bases complementary to DNA strands. • Elongation step continues where the polymerase adds dNTP's from 5' to 3', reading the template from 3' to 5' side, bases are added complementary to the template. • Now first cycle is over and next cycle is continued ,as PCR machine is automated thermocycler the same cycle is repeated up to 30-40 times.
- 15. Steps Involved
- 16. Steps Involved
- 17. Steps Involved
- 18. New Automated PCR OLD PCR
- 19. Optimal PCR Factors PCR Primers DNA Polymerase Annealing Temperature Melting Temperature G/C content
- 20. Optimal PCR Factors PCR Primers correctly designed pair of primers is required primer dimer,hairpin formation should be prevented length of primer DNA Polymerase Annealing Temperature Melting Temperature G/C content
- 21. Optimal PCR Factors PCR Primers DNA Polymerase Thermus aquaticus (Taq - 170°F) Taq polymerase is heat resistant It lacks proof reading exonuclease activity Other polymerases can be used, eg: Tma DNA Polymerase from Thermotoga maritama, Pfu DNA Polymerase from Pyrococcus furiosus. Annealing Temperature Melting Temperature G/C content
- 22. Optimal PCR Factors PCR Primers DNA Polymerase Annealing Temperature Very important since the success and specificity of PCR depend on it because DNA-DNA hybridization is a temperature dependent process. If annealing temperature is too high, pairing between primer and template DNA will not take place then PCR will fail. Ideal Annealing temperature must be low enough to enable hybridization between primer and template but high enough to prevent amplification of nontarget sites. Should be usually 1-2°C or 5°C lower than melting temperature of the template-primer duplex Melting Temperature G/C content
- 23. Optimal PCR Factors PCR Primers DNA Polymerase Annealing Temperature Melting Temperature Temperature at which 2 strands of the duplex dissociate. It can be determined experimentally or calculated from formula Tm = (4(G+C)) + (2(A+T)) G/C content
- 24. Optimal PCR Factors PCR Primers DNA Polymerase Annealing Temperature Melting Temperature G/C content Ideally a primer should have a near random mix of nucleotides, a 50% GC content There should be no PolyG or PolyC stretches that can promote non-specific annealing
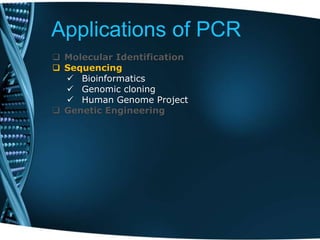
- 25. Applications of PCR Molecular Identification Sequencing Genetic Engineering
- 26. Applications of PCR Molecular Identification Molecular Archaeology Molecular Epidemiology Molecular Ecology DNA fingerprinting Classification of organisms Genotyping Pre-natal diagnosis Mutation screening Drug discovery Genetic matching Detection of pathogens Sequencing Genetic Engineering
- 27. Applications of PCR Molecular Identification Sequencing Bioinformatics Genomic cloning Human Genome Project Genetic Engineering
- 28. Applications of PCR Molecular Identification Sequencing Genetic Engineering Site-directed mutagenesis Gene expression studies
- 29. Variations of PCR PCR is highly versatile technique and has been modified in variety of way to suit specific applications.
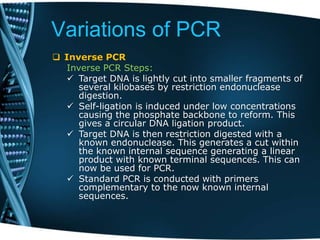
- 30. Variations of PCR Inverse PCR In this method amplification of DNA of unknown sequence is carried out from known sequence. This is especially useful in identifying flanking sequences of various genomic inserts. The inverse PCR method includes a series of digestions and self-ligations with the DNA being cut by a restriction endonuclease. This cut results in a known sequence at either end of unknown sequences. Inverse PCR uses standard PCR however it has the primers oriented in the reverse direction of the usual orientation. The template for the reverse primers is a restriction fragment that has been ligated upon itself to form a circle.
- 31. Variations of PCR Inverse PCR Inverse PCR Steps: Target DNA is lightly cut into smaller fragments of several kilobases by restriction endonuclease digestion. Self-ligation is induced under low concentrations causing the phosphate backbone to reform. This gives a circular DNA ligation product. Target DNA is then restriction digested with a known endonuclease. This generates a cut within the known internal sequence generating a linear product with known terminal sequences. This can now be used for PCR. Standard PCR is conducted with primers complementary to the now known internal sequences.
- 32. Variations of PCR Inverse PCR
- 33. Variations of PCR Ligation-Mediated PCR (LM-PCR) Ligation-mediated PCR uses small DNA oligonucleotide 'linkers' (or adaptors) that are first ligated to fragments of the target DNA. PCR primers that anneal to the linker sequences are then used to amplify the target fragments. This method is deployed for DNA sequencing, genome walking, and DNA foot-printing. The principle of Ligation Mediated PCR (LM-PCR). 1-Ligation with excess of primers, 2-Polymerase chain reaction of individual fragments.
- 34. Variations of PCR Ligation-Mediated PCR (LM-PCR) In LM-PCR, each fragment is amplified independently so that due to intrinsic differences among individual fragments, some fragments are amplified less efficiently than others. This results in non-uniform representation of original genetic material in the resultant amplicon, which consequently leads to loss of genetic information and inaccurate results. Primer-extension step (Step 3): a gene-specific primer (Primer 1) was annealed at 48°C and the primer was extended with Sequenase enzyme at 48°C. Ligation step (Step 4): all extended DNA fragments with a blunt-end and 5'-phosphate group were ligated to an unphosphorylated synthetic asymmetric double-strand linker.
- 35. Variations of PCR Ligation-Mediated PCR (LM-PCR) Linear amplification step (Step 5): a second gene-specific primer (Primer 2) was annealed to DNA fragments for a one-cycle extension using Taq DNA polymerase. Exponential amplification step (Step 6): the primer 2 and the linker primer (the longest of the two oligonucleotides of the linker) were used to exponentially and specifically amplify DNA fragments. Sequencing gel electrophoresis and electroblotting (Step 7): amplified DNA fragments were size-separated on a denaturing 8% polyacrylamide gel and transferred onto a nylon membrane by electroblotting. Hybridization (Step 8): the nylon membrane was hybridized overnight with a gene-specific probe.
- 36. Variations of PCR Ligation-Mediated PCR (LM-PCR) Uses: Is the most sensitive sequencing technique available to map single-stranded DNA breaks at the nucleotide level of resolution using genomic DNA. LM-PCR has been adapted to map DNA damage and reveal DNA–protein interactions inside living cells. However, the sequence context (GC content), the global break frequency and the current combination of DNA polymerases used in LM-PCR affect the quality of the results.
- 37. Variations of PCR Ligation-Mediated PCR (LM-PCR)
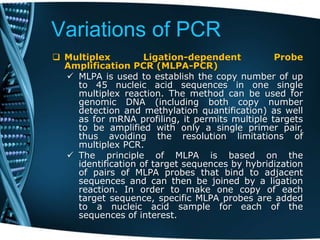
- 38. Variations of PCR Multiplex Ligation-dependent Probe Amplification PCR (MLPA-PCR) MLPA is used to establish the copy number of up to 45 nucleic acid sequences in one single multiplex reaction. The method can be used for genomic DNA (including both copy number detection and methylation quantification) as well as for mRNA profiling, it permits multiple targets to be amplified with only a single primer pair, thus avoiding the resolution limitations of multiplex PCR. The principle of MLPA is based on the identification of target sequences by hybridization of pairs of MLPA probes that bind to adjacent sequences and can then be joined by a ligation reaction. In order to make one copy of each target sequence, specific MLPA probes are added to a nucleic acid sample for each of the sequences of interest.
- 39. Variations of PCR Multiplex Ligation-dependent Probe Amplification PCR (MLPA-PCR) The sequences are then simultaneously amplified with the use of only one primer pair, resulting in a mixture of amplification products, in which each PCR product of each MLPA probe has a unique length. One PCR primer is fluorescently or isotopically labelled so that the MLPA reaction products can be visualized when electrophoresed on a capillary sequencer or a gel. Resulting chromatograms show size-separated fragments ranging from 130 to 490 bp. The peak area or peak height of each amplification product reflects the relative copy number of that target sequence. Comparison of the electrophoresis profile of the tested sample to that obtained with a control sample enables the detection of deletions or duplications of genomic regions of interest
- 40. Variations of PCR Multiplex Ligation-dependent Probe Amplification PCR (MLPA-PCR)
- 41. Variations of PCR Multiplex Ligation-dependent Probe Amplification PCR (MLPA-PCR)
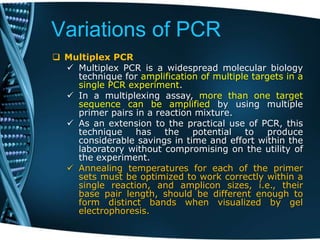
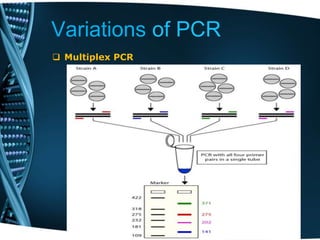
- 42. Variations of PCR Multiplex PCR Multiplex PCR is a widespread molecular biology technique for amplification of multiple targets in a single PCR experiment. In a multiplexing assay, more than one target sequence can be amplified by using multiple primer pairs in a reaction mixture. As an extension to the practical use of PCR, this technique has the potential to produce considerable savings in time and effort within the laboratory without compromising on the utility of the experiment. Annealing temperatures for each of the primer sets must be optimized to work correctly within a single reaction, and amplicon sizes, i.e., their base pair length, should be different enough to form distinct bands when visualized by gel electrophoresis.
- 43. Variations of PCR Multiplex PCR Types of Multiplex PCR: 1. Single template PCR reaction; this technique uses a single template which can be a genomic DNA along with several pairs of forward and reverse primers to amplify specific regions within a template 2. Multiple template PCR reaction; this technique uses multiple templates and several primer sets in the same reaction tube. Presence of multiple primer may lead to cross hybridization with each other and the possibility of mis-priming with other templates.
- 44. Variations of PCR Multiplex PCR Primer Design Parameters for Multiplex PCR: Design of specific primer sets is essential for a successful multiplex reaction. The important primer design considerations described below are a key to specific amplification with high yield. • Primer Length: Multiplex PCR assays involve designing of large number of primers, hence it is required that the designed primer should be of appropriate length. Usually, primers of short length, in the range of 18-22 bases are used. • Melting Temperature: Primers with similar Tm, preferably between 55°C-60°C are used. For sequences with high GC content, primers with a higher Tm (preferably 75°C-80°C) are recommended. A Tm variation of between 3°-5° C is acceptable for primers used in a pool. • Specificity: It is important to consider the specificity of designed primers to the target sequences, while preparing a multiplex assay, especially since competition exists when multiple target sequences are in a single reaction vessel. • Avoid Primer Dimer Formation: The designed primers should be checked for formation of primer dimers, with all the primers present in the reaction mixture. Dimerization leads to unspecific amplification.
- 45. Variations of PCR Multiplex PCR
- 46. Variations of PCR Multiplex PCR Advantages of Multiplex PCR: 1. Internal Controls: Potential problems in a simple PCR include false negatives due to reaction failure or false positives due to contamination. False negatives are often revealed in multiplex assays because each amplicon provides an internal control for the other amplified fragments. 2. Efficiency: The expense of reagents and preparation time is less in multiplex PCR than in systems where several tubes of muniplex PCRs are used. A multiplex reaction is ideal for conserving costly polymerase and templates in short supply. 3. Indication of Template Quality: The quality of the template may be determined more effectively in multiplex than in a simple PCR reaction. 4. Indication of Template Quantity: The exponential amplification and internal standards of multiplex PCR can be used to assess the amount of a particular template in a sample. To quantitate templates accurately by multiplex PCR, the amount of reference template, the number of reaction cycles, and the minimum inhibition of the theoretical doubling of product for each cycle must be accounted.
- 47. Variations of PCR Multiplex PCR Uses of Multiplex PCR: Its has been found useful in: Pathogen Identification, High Throughput SNP Genotyping, Mutation Analysis, Gene Deletion Analysis, Template Quantification, Linkage Analysis, RNA Detection, Forensic Studies.
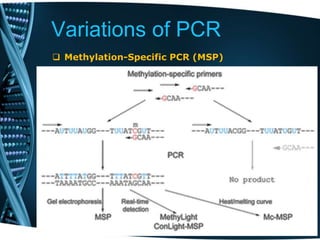
- 48. Variations of PCR Methylation-Specific PCR (MSP) • Methylation-specific PCR (MSP) is used to identify patterns of DNA methylation at cytosine-guanine (CpG) islands in genomic DNA . • Target DNA is first treated with sodium bisulphite, which converts unmethylated cytosine bases to uracil, which is complementary to adenosine in PCR primers. • Two amplifications are then carried out on the bisulphite-treated DNA: One primer set anneals to DNA with cytosines (corresponding to methylated cytosine), and the other set anneals to DNA with uracil (corresponding to unmethylated cytosine). • MSP used in Q-PCR provides quantitative information about the methylation state of a given CpG island.
- 49. Variations of PCR Methylation-Specific PCR (MSP) • Treatment of DNA with bisulphite converts cytosine residues to uracil, but leaves 5-methylcytosine residues unaffected. • Thus, bisulphite treatment introduces specific changes in the DNA sequence that depend on the methylation status of individual cytosine residues, yielding single- nucleotide resolution information about the methylation status of a segment of DNA. • The objective of this analysis is therefore reduced to differentiating between single nucleotide polymorphisms (cytosines and thymidine) resulting from bisulphite conversion.
- 50. Variations of PCR Methylation-Specific PCR (MSP) • The MethyLight method is based on MSP, but provides a quantitative analysis using real-time PCR. • Methylated-specific primers are used, and a methylated-specific fluorescence reporter probe is also used that anneals to the amplified region. • In alternative fashion, the primers or probe can be designed without methylation specificity if discrimination is needed between the CpG pairs within the involved sequences. • Quantitation is made in reference to a methylated reference DNA. A modification to this protocol to increase the specificity of the PCR for successfully bisulphite-converted DNA (ConLight-MSP) uses an additional probe to bisulphite-unconverted DNA to quantify this non-specific amplification.
- 51. Variations of PCR Methylation-Specific PCR (MSP)
- 52. Variations of PCR Hot Start PCR This is a technique that reduces non-specific amplification during the initial set up stages of the PCR The technique may be performed manually by heating the reaction components to the melting temperature (e.g., 95°C) before adding the polymerase Specialized enzyme systems have been developed that inhibit the polymerase's activity at ambient temperature, either by the binding of an antibody or by the presence of covalently bound inhibitors that only dissociate after a high- temperature activation step DNA Polymerase- Eubacterial type I DNA polymerase, Pfu These thermophilic DNA polymerases show a very small polymerase activity at room temperature.
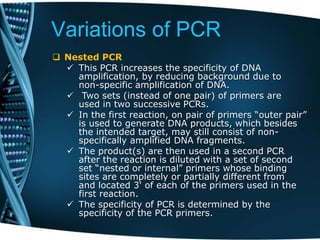
- 53. Variations of PCR Nested PCR This PCR increases the specificity of DNA amplification, by reducing background due to non-specific amplification of DNA. Two sets (instead of one pair) of primers are used in two successive PCRs. In the first reaction, on pair of primers “outer pair” is used to generate DNA products, which besides the intended target, may still consist of non- specifically amplified DNA fragments. The product(s) are then used in a second PCR after the reaction is diluted with a set of second set “nested or internal” primers whose binding sites are completely or partially different from and located 3' of each of the primers used in the first reaction. The specificity of PCR is determined by the specificity of the PCR primers.
- 54. Variations of PCR Nested PCR For example, if your primers bind to more than one locus (e.g. paralog or common domain), then more than one segment of DNA will be amplified. To control for these possibilities, investigators often employ nested primers to ensure specificity. Nested PCR means that two pairs of PCR primers were used for a single locus. The first pair amplified the locus as seen in any PCR experiment. The second pair of primers (nested primers) bind within the first PCR product and produce a second PCR product that will be shorter than the first one. The logic behind this strategy is that if the wrong locus were amplified by mistake, the probability is very low that it would also be amplified a second time by a second pair of primers.
- 55. Variations of PCR Nested PCR
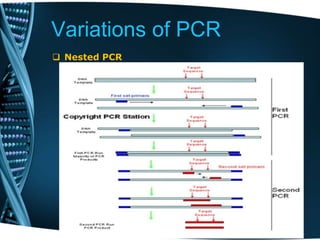
- 56. Variations of PCR Nested PCR Nested PCR strategy: Segment of DNA with dots representing non- discript DNA sequence of unspecified length. The double lines represent a large distance between the portion of DNA illustrated in this figure. The portions of DNA shown with four bases in a row represent PCR primer binding sites, though real primers would be longer. The first pair of PCR primers (blue with arrows) bind to the outer pair of primer binding sites and amplify all the DNA in between these two sites
- 57. Variations of PCR Nested PCR Nested PCR strategy: PCR product after the first round of amplificaiton. Notice that the bases outside the PCR primer pair are not present in the product. PCR product after the first round of amplificaiton. Notice that the bases outside the PCR primer pair are not present in the product.
- 58. Variations of PCR Nested PCR Nested PCR strategy: Final PCR product after second round of PCR. The length of the product is defined by the location of the internal primer binding sites.
- 59. Variations of PCR Nested PCR Uses of Nested PCR: When a complete genome sequence is known, it is easier to be sure you will not amplify the wrong locus but since very few of the world's genomes have been sequenced completely, nested primers will continue to be an important control for many experiments.
- 60. Variations of PCR AFLP PCR AFLP is a highly sensitive PCR-based method for detecting polymorphisms in DNA. AFLP can be also used for genotyping individuals for a large number of loci Genomic DNA is digested with one or more restriction enzymes. tetracutter (MseI) and a hexacutter (EcoRI) Ligation of linkers to all restriction fragments Pre-selective PCR is performed using primers which match the linkers and restriction site specific sequences Electrophoretic separation and amplicons on a gel matrix, followed by visualisation of the band pattern
- 61. Variations of PCR AFLP PCR
- 62. Variations of PCR Anchored PCR A small sequence of nucleotides can be attached or tagged to target DNA. The anchor is frequently a poly G to which a poly C primer is used.
- 63. Variations of PCR Anchored PCR
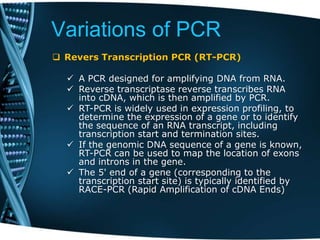
- 64. Variations of PCR Revers Transcription PCR (RT-PCR) A PCR designed for amplifying DNA from RNA. Reverse transcriptase reverse transcribes RNA into cDNA, which is then amplified by PCR. RT-PCR is widely used in expression profiling, to determine the expression of a gene or to identify the sequence of an RNA transcript, including transcription start and termination sites. If the genomic DNA sequence of a gene is known, RT-PCR can be used to map the location of exons and introns in the gene. The 5' end of a gene (corresponding to the transcription start site) is typically identified by RACE-PCR (Rapid Amplification of cDNA Ends)
- 65. Variations of PCR Revers Transcription PCR (RT-PCR)
- 66. Variations of PCR Revers Transcription PCR (RT-PCR)
- 67. Variations of PCR RACE PCR Used to obtain 3' and 5' end sequence of cDNA transcripts.
- 68. Variations of PCR RACE PCR
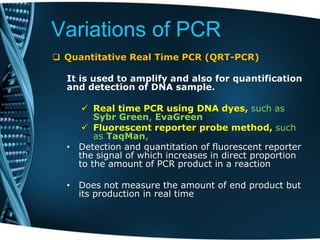
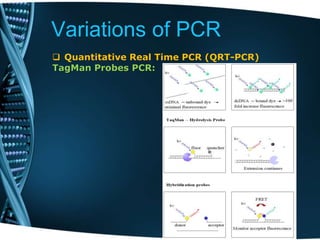
- 69. Variations of PCR Quantitative Real Time PCR (QRT-PCR) It is used to amplify and also for quantification and detection of DNA sample. Real time PCR using DNA dyes, such as Sybr Green, EvaGreen Fluorescent reporter probe method, such as TaqMan, • Detection and quantitation of fluorescent reporter the signal of which increases in direct proportion to the amount of PCR product in a reaction • Does not measure the amount of end product but its production in real time
- 70. Variations of PCR Quantitative Real Time PCR (QRT-PCR)
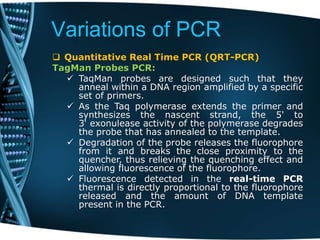
- 71. Variations of PCR Quantitative Real Time PCR (QRT-PCR) TagMan Probes PCR: TaqMan probes are designed such that they anneal within a DNA region amplified by a specific set of primers. As the Taq polymerase extends the primer and synthesizes the nascent strand, the 5' to 3‘ exonulease activity of the polymerase degrades the probe that has annealed to the template. Degradation of the probe releases the fluorophore from it and breaks the close proximity to the quencher, thus relieving the quenching effect and allowing fluorescence of the fluorophore. Fluorescence detected in the real-time PCR thermal is directly proportional to the fluorophore released and the amount of DNA template present in the PCR.
- 72. Variations of PCR Quantitative Real Time PCR (QRT-PCR) TagMan Probes PCR:
- 73. Variations of PCR Quantitative Real Time PCR (QRT-PCR) TagMan Probes PCR:
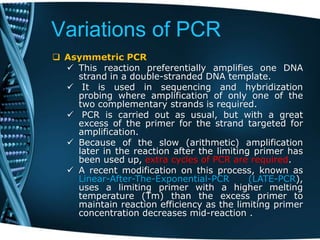
- 74. Variations of PCR Asymmetric PCR This reaction preferentially amplifies one DNA strand in a double-stranded DNA template. It is used in sequencing and hybridization probing where amplification of only one of the two complementary strands is required. PCR is carried out as usual, but with a great excess of the primer for the strand targeted for amplification. Because of the slow (arithmetic) amplification later in the reaction after the limiting primer has been used up, extra cycles of PCR are required. A recent modification on this process, known as Linear-After-The-Exponential-PCR (LATE-PCR), uses a limiting primer with a higher melting temperature (Tm) than the excess primer to maintain reaction efficiency as the limiting primer concentration decreases mid-reaction .
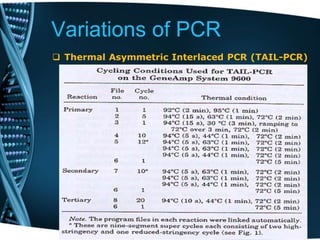
- 75. Variations of PCR Thermal Asymmetric Interlaced PCR (TAIL-PCR) This reaction is applied in the isolation of an unknown sequence flanking a known sequence. Within the known sequence, TAIL-PCR uses a nested pair of primers with differing annealing temperatures; a degenerate primer is used to amplify in the other direction from the unknown sequence . Uses: TAIL-PCR as a powerful tool for amplifying insert end segments from P1, BAC and YAC clones, the amplified products were highly specific and suitable as probes for library screening and as templates for direct sequencing while the recover insert ends can also be used for chromosome walking and mapping.
- 76. Variations of PCR Thermal Asymmetric Interlaced PCR (TAIL-PCR) • Nested, insertion-specific primers are used together with arbitrary degenerate primers (AD primers), which are designed to differ in their annealing temperatures. • Alternating cycles of high and low annealing temperature yield specific products bordered by an insertion-specific primer on one side and an AD primer on the other. • Further specificity is obtained through subsequent rounds of TAIL-PCR, using nested insertion-specific primers. • The increasing availability of whole genome sequences renders TAIL-PCR an attractive tool to easily identify insertion sites in large genome tagging populations through the direct sequencing of TAIL-PCR products. • For large-scale functional genomics approaches, it is desirable to obtain flanking sequences for each individual in the population in a fast and cost- effective manner.
- 77. Variations of PCR Thermal Asymmetric Interlaced PCR (TAIL-PCR)
- 78. Variations of PCR Thermal Asymmetric Interlaced PCR (TAIL-PCR)
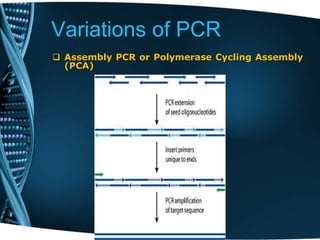
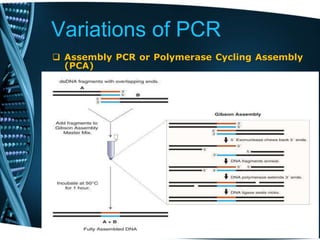
- 79. Variations of PCR Assembly PCR or Polymerase Cycling Assembly (PCA) This entails the artificial synthesis of long DNA sequences by performing PCR on a pool of long oligonucleotides with short overlapping segments. The oligonucleotides alternate between sense and antisense directions, and the overlapping segments determine the order of the PCR fragments, thereby selectively producing the final long DNA product.
- 80. Variations of PCR Assembly PCR or Polymerase Cycling Assembly (PCA)
- 81. Variations of PCR Assembly PCR or Polymerase Cycling Assembly (PCA)
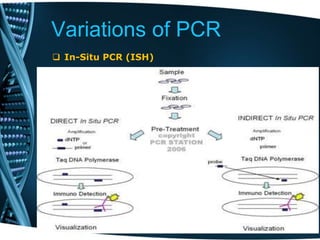
- 82. Variations of PCR In-Situ PCR (ISH) A polymerase chain reaction that actually takes place inside the cell on a slide. In situ PCR amplification can be performed on fixed tissue or cells. Uses: Detection and diagnosis of viruses and other infectious agents in specific cell types within tissues. Detection and characterization of tumor cells within tissue. Detection and diagnosis of genetic mutations in inherited diseases. Detection of gene and gene expression in a tissue. Any assay in which identification of cells expressing a target gene is required. Main advantages are low background, high specificity, fast assay with shorter turn-around time and no need of radioactive chemicals.
- 83. Variations of PCR In-Situ PCR (ISH)
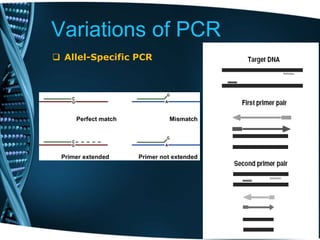
- 84. Variations of PCR Allel-Specific PCR Selective PCR amplification of the alleles to detect single nucleotide polymorphism (SNP) Selective amplification is usually achieved by designing a primer such that the primer will match or mismatch one of the alleles at the 3’ end of the primer.
- 85. Variations of PCR Allel-Specific PCR
- 86. Variations of PCR Single Cell PCR It is now possible to amplify and examine minute quantities of rare genetic material, the limit of this exploration being the single cell. Single cell PCR has applications in many areas, and has great application especially in the field of prenatal diagnostics. In prenatal diagnosis, single cell PCR has made possible preimplantation genetic analysis and the use of fetal cells enriched from the blood of pregnant women for the assessment of single- gene Mendelian disorders. Single-cell PCR has not only proven its usefulness in diagnostics, but also lately has been very useful to basic scientists investigating immunological, neurological and developmental problems.
- 87. Variations of PCR Helicase-Dependent Amplification This PCR is similar to traditional PCR, but uses a constant temperature rather than cycling through denaturation and annealing/extension cycles. DNA helicase, an enzyme that unwinds DNA, is used in place of thermal denaturation. Alu PCR The pcr is performed using Alu primers designed to have recognition sequence of Alu restriction enzyme. Used as a method of obtaining a fingerprint of bands from an uncharacterized human DNA.
- 88. Variations of PCR LONG PCR Long PCR is a PCR is which extended or longer than standard PCR, meaning over 5 kilobases (frequently over 10 kb). Long PCR is usually only useful if it is accurate. Thus, special mixtures of proficient polymerases along with accurate polymerases such as Pfu are often mixed together. Applications of Long PCR: Long PCR is often used to clone larger genes or large segments of DNA which standard PCR cannot.
- 89. Variations of PCR Arbitrarily Primed PCR (AP-PCR) Arbitrarily Primed PCR (AP-PCR) or Random Amplified Polymorphic DNA (RAPD) are methods of creating genomic fingerprints from species of which little is known about target sequence to be amplified. TAP-PCR AP-PCR run at three different annealing temperature
- 90. Variations of PCR Colony PCR The screening of bacterial (E.Coli) or yeast clones for correct ligation or plasmid products. Selected colonies of bacteria or yeast are picked with a sterile toothpick or pipette tip from a growth (agarose) plate. This is then inserted into the PCR master mix or pre-inserted into autoclaved water. PCR is then conducted to determine if the colony contains the DNA fragment or plasmid of interest.
- 91. Variations of PCR LAMP (Loop-Mediated isothermal amplification) Assay: It is a Modified type of the PCR using 3-6 primers sets one of them is loop like primer. This test use Bst-polymerase (Bacillus stearothermophilus DNA Polymerase) enzyme. Using only two temperatures (63°C and 85°C for one hour), may be carry out in water bath.
- 92. Variations of PCR The Digital PCR The Digital polymerase chain reaction simultaneously amplifies thousands of samples, each in a separate droplet within an emulsion . Overlap-Extention PCR A genetic engineering technique allowing the construction of a DNA sequence with an alteration inserted beyond the limit of the longest practical primer length . Solid Phase PCR Encompasses multiple meanings, including Colony Amplification (where PCR colonies are derived in a gel matrix, for example), 'Bridge PCR' (primers are covalently linked to a solid-support surface), conventional Solid Phase PCR (where Asymmetric PCR is applied in the presence of solid support bearing primer with sequence matching one of the aqueous primers) and Enhanced Solid Phase PCR (where conventional Solid Phase PCR can be improved by employing high Tm and nested solid support primer with optional application of a thermal 'step' to favour solid support priming)
- 93. Variations of PCR Box PCR Box elements are repetitive sequence elements in bacterial genome such as Streptococcus genome. Single primer targeting to the repeats can be used to fingerprint bacterial species. Competitive PCR(cPCR) This is a method used for quantifying DNA using real time PCR. A competitor internal standard is co amplified with the target DNA and the target is quantified from the melting curves of the target and its competitor. Consensus PCR This PCR is carried out by using flanking primers to amplify repeat regions from a no. of species. In this case degenerate/consensus primers can be used for amplifying the flanking sequences.
- 94. Variations of PCR Degenerate PCR In this instead of using specific PCR primers for a given sequence, mixed PCR primers will be used. That is “wobbles” are inserted into the primers in case if the exact sequence of gene is not known, so that there will be more than one possibility for exact amplifications. Degenerate PCR has proven to be a powerful tool to find ‘new’ gene or gene families. By aligning the sequences from a no. of related proteins the conserved and variables part can be determined. Based on this information one can use conserved protein motifs for starting points for designing degenerate PCR primers. Degenerate oligonucleotide-primed PCR(DOR PCR) PCR amplification of limited sample by using degenerate PCR primers is called DOR PCR.
- 95. Variations of PCR Differential Display PCR (DD PCR) It is used for cloning purpose; it combines the comparative analysis of several samples with the sensitivity of PCR. Forensic PCR The VNTR locus is PCR amplified to compare DNA samples from different sources. Hairpin PCR A method for error free DNA amplification for mutation detection. It first converts a DNA sequence to a hairpin. True mutations will maintain hairpin structure during amplification while PCR errors will disrupt the hairpin structure. PCR ELISA The PCR products are labeled(digoxigenin) during amplification. A capture probe specific to PCR amplicon is used to immobilize the amplicon to immune-well plate. ELISA is then used against the label(anti- digoxigenin) to quantitate PCR products.
- 96. Variations of PCR Touchdown PCR (Step-Down PCR) A variant of PCR that aims to reduce nonspecific background by gradually lowering the annealing temperature as PCR cycling progresses. The annealing temperature at the initial cycles is usually a few degrees (3-5°C) above the Tm of the primers used, while at the later cycles, it is a few degrees (3-5°C) below the primer Tm. The higher temperatures give greater specificity for primer binding, and the lower temperatures permit more efficient amplification from the specific products formed during the initial cycles. Miniprimer PCR This reaction uses a thermostable polymerase (S-Tbr) that can extend from short primers ("smalligos") as short as 9 or 10 nucleotides. This method permits PCR targeting to smaller primer binding regions, and is used to amplify conserved DNA sequences, such as the 16S (or eukaryotic 18S) rRNA gene.
- 97. Variations of PCR Rep-PCR Is used for Genomic Fingerprinting of plant- associated bacteria and computer-assisted plant analyses. The genomic fingerprinting method employed is based on the use of DNA primers corresponding to naturally occurring interspersed repetitive elements in bacteria such as REP,ERIC and BOX elements. Vectorette-PCR This method enables the amplification of specific DNA fragments in situation where sequence of only one primer is known. Thus it extends the application of PCR to the stretches of DNA where the sequence information is only available at one end.
- 98. Variations of PCR Universal Fast Walking PCR Used for genome walking and genetic fingerprinting using a more specific 'two-sided' PCR than conventional 'one- sided' approaches (using only one gene-specific primer and one general primer - which can lead to artefactual 'noise') by virtue of a mechanism involving lariat structure formation. Streamlined derivatives of UFW are LaNe RAGE (lariat-dependent nested PCR for rapid amplification of genomic DNA ends), 5'RACE LaNe and 3'RACE LaNe . Variable Number of Tandem Repeats (VNTR) PCR This method targets areas of the genome that exhibit length variation. The analysis of the genotypes of the sample usually involves sizing of the amplification products by gel electrophoresis. Analysis of smaller VNTR segments known as Short Tandem Repeats (or STRs) is the basis for DNA Fingerprinting databases such as CODIS .
- 99. Variations of PCR Intersequence-Specific PCR (ISSR-PCR) This is a method for DNA fingerprinting that uses primers selected from segments repeated throughout a genome to produce a unique fingerprint of amplified product lengths. The use of primers from a commonly repeated segment is called Alu-PCR, and can help amplify sequences adjacent (or between) these repeats.
- 100. Variations of PCR Other types of PCR Overlap extension PCR Solid phase PCR and so on…………………..
- 101. Comparison PCR & Cloning Parameter PCR Gene cloning 1. Final result Selective amplification of specific sequence Selective amplification of specific sequence 2. Manipulation In vitro In vitro and in vivo 3. Selectivity of the specific segment from complex DNA First step Last step 4. Quantity of starting material Nanogram (ng) Microgram (m) 5. Biological reagents required DNA polymerase (Taq polymerase) Restriction enzymes, Ligase, vector. bacteria 6. Automation Yes No 7. Labour intensive No Yes 8. Error probability Less More 9. Applications More Less 10. Cost Less More 11. User’s skill Not required Required 12. Time for a typical experiment Four hours Two to four days
- 102. Advantages of PCR PCR in clinical diagnosis PCR in DNA sequencing PCR in Forsenic Medicine PCR in Gene manipulation and expression studies PCR in comparative study of genomics PCR in comparison with gene cloning
- 103. Limitations of PCR Sequence Information Amplicon size Error rate during amplification Sensitivity to inhibitors Contamination Artefacts
- 104. Part II: Thermostable DNA Polymerases
- 105. Contents of part II: • Discovery • Properties • Taq DNA Polymerase • Other Thermostable Polymerases • The Error Rate • Reliability / Fidelity
- 106. Discovery • The original report of this enzyme, purified from the hot springs bacterium Thermus aquaticus, was published in 1976. • Roughly 10 years later, the polymerase chain reaction was developed and shortly thereafter "Taq" became a household word in molecular biology circles. • *THE DARNDEST PLACES: Scientists isolated the thermostable DNA polymerase Taq, an enzyme that drives PCR, from Thermus aquaticus Yellowstone type-1, a resident of geysers like this one at Yellowstone National Park.
- 107. Properties • The thermophilic DNA polymerases, like other DNA polymerases, catalyze template-directed synthesis of DNA from nucleotide triphosphates. • A primer having a free 3‘ hydroxyl is required to initiate synthesis • Magnesium ion is necessary. • In general, they have maximal catalytic activity at 75 to 80℃, and substantially reduced activities at lower temperatures. • At 37℃, Taq polymerase has only about 10% of its maximal activity.
- 108. Taq DNA Polymerase • Recombinant Taq DNA Polymerase is the enzyme of choice for most PCR applications. • The half-life of enzyme is >40 minutes at 95°C. • The error rate of Taq DNA Polymerase in PCR is 2.2x10-5 errors per nt per cycle;
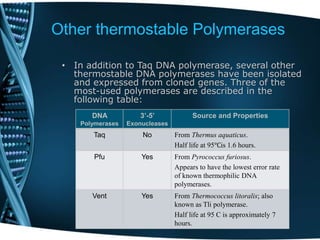
- 109. Other thermostable Polymerases • In addition to Taq DNA polymerase, several other thermostable DNA polymerases have been isolated and expressed from cloned genes. Three of the most-used polymerases are described in the following table: Source and Properties3’-5’ Exonucleases DNA Polymerases From Thermus aquaticus. Half life at 95℃is 1.6 hours. NoTaq From Pyrococcus furiosus. Appears to have the lowest error rate of known thermophilic DNA polymerases. YesPfu From Thermococcus litoralis; also known as Tli polymerase. Half life at 95 C is approximately 7 hours. YesVent
- 110. The Error Rate • One of the most discussed characteristics of thermostable polymerases is their error rate. • Error rates are measured using several different assays, and as a result, estimates of error rate vary, particularly when the assays are performed by different labs. The Total Error RateDNA Polymerases 1 x 10-4 to 2 x 10-5 errors per base pairTaq appears to have the lowest error rate at roughly 1.5 x 10-6 error per base pair Pfu between Taq and PfuVent
- 111. Reliability/Fidelity Average error rates(mutation frequency/bp/duplication) increased as follows: Pfu (1.3 x 10-6) Deep Vent (2.7 x 10-6) Vent (2.8 x 10-6) Taq (8.0 x 10-6) exo- Pfu and UlTma (approximately 50 x 10-6)
- 112. Reference • Yasumasa Kimura et al. Optimization of turn-back primers in isothermal amplification (http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3089485/ • R. Manojkumar and Mrudula Varanat(2006) Polymerase Chain Reaction: Types and Its Application in theField of Biology. International journal of tropical medicine 1 (4):156-161 • Voet,D, Voet,J. Biochemistry Vol.1 3rd ed. • Alberts, Johnson, Lewis. Molecular Biology of The Cell 4th ed. • Introduction to Plant Biotechnology By- H.S. Chawala http://arbl.cvmbs.colostate.edu/hbooks/genetics/biotech/enzymes/hotpolys .html http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=146123&rendert ype=abstract http://www.fermentas.com/techinfo/pcr/dnaamplprotocol.htm http://www.fermentas.com/techinfo/pcr/pcrprotocolpfu.htm