Preprint
Review
Personalized Plasma Medicine for Cancer: Transforming Treatment Strategies with Mathematical Modeling and AI Techniques
Altmetrics
Downloads
313
Views
184
Comments
1
This version is not peer-reviewed
Submitted:
22 September 2023
Posted:
25 September 2023
You are already at the latest version
Alerts
Abstract
Plasma technology shows tremendous potential for revolutionizing oncology research and treatment. Reactive oxygen and nitrogen species, electromagnetic emissions generated through gas plasma jets, have attracted significant attention due to their selective cytotoxicity towards cancer cells. To leverage the full potential of plasma medicine, researchers have explored the use of mathematical models and various subsets of machine learning, such as reinforcement learning, and deep learning. This review emphasizes the significant application of AI algorithms in the adaptive plasma system, paving the way for precision and dynamic cancer treatment. Realizing the full potential of AI in plasma medicine, requires research efforts, data sharing and interdisciplinary collaborations. Unravelling the complex mechanisms, developing real-time diagnostics, and optimizing AI models will be crucial to harness the true power of plasma technology in oncology. The integration of personalized and dynamic plasma therapies, alongside AI and diagnostic sensors, presents a transformative approach to cancer treatment with the potential to improve outcomes globally.

Keywords:
Subject: Biology and Life Sciences - Life Sciences
1. Introduction
Plasma technology, rapidly emerging field at the intersection of plasma physics and medicine holds immense potential for biomedical applications [1-9]. Plasma, known as the fourth state of matter is typically formed by heating gas such as inert gases like argon or helium, or air. When heated, the high-speed electrons collide with atoms or molecules causing the removal of electrons and generating highly reactive positive and negative ions. These ions are conductive and can oscillate in response to electric and magnetic fields. The oscillations can give rise to emission of various electromagnetic waves [10]. Additionally, there are also low energy electrons that can only excite particles without causing ionization. These excited particles release energy in the form of photons. Plasma or ionized gas, is a complex mixture comprising electrons, positively and negatively charged ions, radicals, neutral atoms and molecules, atoms at excited state, electromagnetic waves, and electromagnetic fields [11]. The degree of ionization and the specific composition of plasma is dependent on factors such as temperature, pressure, energy supply, nature of the gas, electron density ranges, the devise used and operational conditions utilized to generate the plasma [12,13]. The most common type of low temperature atmospheric pressure plasma is the cold atmospheric plasma jets (CAPJ) has been well established in the field of biology and medicine [14].
Atmospheric plasma, can self-organize into different patterns with modified plasma compositions in response to various operational parameters and interactions with the target environment [15,16]. This transition from spatially homogeneous state to self-organized patterns in response to various parameters like discharge voltage [17-19], electrical permittivity of target cells [20] can play an important role in the therapeutic outcome of plasma treatment for cancer [21,22]. Self-organization might allow an adaptive and self-adaptive plasma system that can dynamically adjust and optimize plasma treatment conditions based on real-time feedback [16]. It aims to maximize the therapeutics on cancer cells while ensuring safety and efficacy.
This review focuses specifically on the recent prominence of cold atmospheric plasma (CAP) in the field of oncology, with special emphasis on the application of various mathematical models and artificial intelligence (AI) techniques. CAP's potential to revolutionize cancer treatment is explored in this review, drawing attention to its unique adaptive and self-adaptive characteristics and the role of mathematical modelling and advanced algorithms in advancing its application in precision medicine.
2. Mathematical modeling
In cancer research, mathematical modeling plays a pivotal role in comprehending the intricate dynamics of cancer in response to various treatments [23,24]. It complements cancer research by providing valuable quantitative predictions to unravel the complexity of cancer and open new avenues for developing effective treatments [25]. Numerous mathematical models, often rooted in ordinary differential equations (ODEs), have explored various aspects of cancer research, including treatment strategies [26-34], immune responses [35-41], treatment sensitivity and resistance [42], the effects of treatment combinations [43-49], habitat dynamics [50], tumor heterogeneity [51,52], and treatment optimization [53]. Their collective contribution lies in furnishing predictions for optimal dosing, treatment regimens, and scheduling, all while minimizing adverse side effects and maximizing therapeutic gains. This avenue holds promise for personalized cancer care, customizing treatments based on individual patient traits and diverse tumor characteristics, including size, type, growth rate, heterogeneity, genetics, and immune composition [37,54]. For mathematical models to operate with precision, the accurate identification of critical parameters hinges on the acquisition of frequent and precise data [55]. ODE-based equations are commonly employed in mechanistic and deterministic models where underlying mechanisms and deterministic relationships are of primary interest. Beyond the realm of ODEs, fractional calculus emerges as a potential frontier for refining cancer modeling, particularly in capturing memory effects and intricate aspects of tumor growth [56-58]. Fractional calculus enables differentiation and integration of non-integer orders, extending beyond traditional calculus. In cancer research, it finds application by modeling anomalous diffusion processes, characterizing tumor behavior more accurately, and incorporating memory effects into models, deepening our understanding of tumor dynamics.
Several mathematical models have been validated through preclinical investigations and clinical trials, demonstrating their potential for shaping effective clinical trial designs [59-65]. Notably, Dean et al. [65] engineered a mathematical model tailored to refine radiation therapy schedules for glioblastoma, a formidable brain cancer. The model recommended an almost-optimal re-irradiation regimen for recurrent glioblastoma patients: 7 days of 3.96 Gy (administered once per day), followed by 9 days of 1.0 Gy (administered thrice per day). Strikingly, this regimen demonstrated both feasibility and safety in all 14 patients (100%) who received it, marking a notable achievement in glioblastoma treatment. Although preliminary results hint at enhanced tumor control, the imperative for larger-scale trials to substantiate its efficacy remains paramount. These findings prove the invaluable role of mathematical modeling in fine-tuning cancer treatment strategies, offering promising prospects for future clinical applications.
After the proposal of adaptive and self-adaptive plasma [66], an exponential growth model was developed [67] to predict the dynamic response of cancer cells (U-87 MG and MDA-MB-231) under CAP treatment [18]. This model successfully derived optimal treatment conditions for two distinct cancer cell lines: U-87 MG and MDA-MB-231. For U-87 MG, the study aimed for a desired ratio of cancer cell viability (rd) of 0.5, leading to optimal treatment parameters of ∆t (Treatment Duration) at 65.1 seconds and U (Plasma Discharge Voltage) at 3.367 kV. For MDA-MB-231, targeting a rd of 0.3 resulted in optimal settings of ∆t at 71.6 seconds and U at 3.315 kV. These specific values were attained through rigorous numerical optimization techniques applied to the mathematical model describing cancer cell responses to CAP exposure. Furthermore, the research extended the optimal control framework to employ an optimal feedback control strategy using Model Predictive Control (MPC). This approach facilitates real-time adaptation of treatment parameters based on actual cancer cell responses, effectively mitigating the impact of uncertainties, and ensuring consistent maintenance of the desired level of cancer cell growth inhibition. The model thus offers a powerful tool for quantitatively optimizing CAP treatment parameters, accounting for dynamic cancer cell responses over time to achieve precise and targeted levels of growth inhibition. However, it is important to note that the model's predictive accuracy is contingent upon the quality and representativeness of the experimental data, necessitating comprehensive validation through experiments. Additionally, the model may not encompass all biological complexities, potentially introducing uncertainties into its predictions.
In recent studies, indirect treatment using plasma treated liquids (PTLs) [68-70] or plasma treated hydrogels (PTHs) [71] has shown enhanced cytotoxicity and anticancer immune responses, attributed to the long-lived reactive oxygen and nitrogen species (RONS) such as H2O2, NO2−, NO3− . These treatments have demonstrated greater effectiveness than direct plasma treatment [72-76]. However, despite their advantages, concerns regarding the lack of consistent molecular-level analysis and precise control over their chemical compositions have been raised, limiting their clinical suitability. Mathematical modeling has been instrumental in investigating the behavior of RONS in cancer treatment with PTLs, allowing for parameter exploration to optimize treatment strategies. Bengston and Bogaerts [77,78] developed a valuable quantitative framework using mathematical modeling to investigate the behavior of reactive oxygen and nitrogen species (RONS) in cancer treatment with PTLs, enabling parameter exploration for treatment optimization. This model suggests that specific cell features, such as the H2O2 membrane diffusion rate constant and intracellular catalase concentration, influence the response to PTLs. It also proposes that certain reactions involving CO3•– (carbonate radical anion) [79] and catalase may be responsible for the enhanced anti-cancer effect observed when combining H2O2 and NO2− in PTLs.
Limitations associated with mathematical models in the context of cold atmospheric plasma (CAP) treatment are multifaceted and have implications for their reliability and applicability. These limitations include a strong reliance on the accuracy of input data, potential oversimplification of complex biological processes, the critical need for experimental validation, a confined scope of parameterization, potential variations across different cell types, assumptions of homogeneity, reliance on literature-derived values, and inherent model uncertainty. Future advancements in this field should focus on developing patient-specific models, refining parameter estimation techniques, exploring the utility of biomarkers, enhancing data collection methods, and conducting rigorous validations. Notably, the integration of AI techniques holds promise in addressing these limitations by improving prediction accuracy, automating feature engineering, accommodating nonlinear relationships, handling noisy and incomplete data, enabling real-time adaptability, and incorporating diverse data types [80]. Mathematical modeling entails the creation of mathematical representations for real-world systems, while AI/ML approaches use data-driven techniques to learn patterns and make predictions, often incorporating mathematical principles, formulas, and statistical methods within their frameworks [81,82]. By combining the strengths of mathematical modeling and AI/ML, we can potentially offer improved patient care and treatment strategies [83], enhancing clinical trials and personalized medicine.
3. AI techniques for Adaptive Plasma system
Artificial intelligence (AI) is about creating “thinking machines” that can make decisions on their own. Machine learning (ML) is the subset of AI that aims at creating “learning machines” in which machines/tools learn from data and perform tasks without explicit programing [84]. A tutorial review on widely used ML methods that can help in discovering patterns and relationships in datasets that may be challenging for human intuition are well described by Bonzanini et al. [85].
The evolution of AI, particularly with the integration of ML, has opened new possibilities in oncology, revolutionizing every facet of cancer care. It has significantly enhanced cancer diagnosis [86-90], prognosis [91-95], and the prediction of metastasis [96-98]. Treatment selection [99], efficacy [100-102] and response assessment [103-107], and outcome prediction [108-112] have also seen remarkable enhancements. These advancements have paved the way for highly personalized cancer treatments, offering a ray of hope within the continually evolving healthcare landscape [113-115]. Although ML has been applied in various treatment approaches in cancer medicine, its use in plasma medicine is relatively limited (Figure 1). The potential of ML in accelerating plasma research towards personalized cancer treatment is well discussed [116-119].
3.1. Reinforcement learning
In machine learning (ML), algorithms are used to train and construct models using large datasets containing relevant information, such as input features (e.g., varying operational parameters of the adaptive plasma system) and target labels (e.g., treatment responses by the cancer cells). The objective is to analyze the datasets, identify patterns and relationships between input features and target labels, and adjust the internal parameters of the algorithm iteratively to improve its predictive accuracy (e.g., optimizing parameters in the adaptive plasma system for selective cytotoxicity towards cancer cells). The goal is to minimize the difference between the model's predictions and the actual outputs during training, making the model "trained" and capable of making accurate predictions.
Different ML approaches such as reinforcement learning (RL) and deep learning (DL) have different methods to update their parameters during training. For example, in RL process, the agent (adaptive plasma system) learns, gets trained and improves its behavior in an environment (cancer cells) through its interaction experience [120]. It takes actions, receives feedback in the form of rewards and penalties based on the effectiveness of its treatment strategies, and uses this feedback to update its internal parameters and improve its decision-making. The system aims to find an optimal strategy that maximizes the cumulative reward it receives.
Hou et al. [116], developed an adaptive plasma framework for CAP cancer treatments using integration of empirical dynamic modeling and reinforcement learning. The empirical dynamic model was constructed using data collected from in vitro experiments [18] that provided information on how cancer cell viability changes over time under various CAP treatment conditions (eg. treatment duration from 0 to 180 seconds, and the discharge voltage between 3.16 kV and 3.71 kV). The empirical data obtained from in vitro experiments may represent specific scenarios and treatment conditions. To estimate for a wide range of treatment scenarios, a Gaussian process, a type of statistical modeling tool, that helps the model to capture the variability and uncertainty inherent in real-world clinical settings was incorporated. The CAP cancer treatment was then modelled as a Markov decision process (MDP) and conjugated with safe Q-learning. The MDP represents the sequence of states and actions in the treatment process. Each state corresponds to a specific situation or condition of the cancer cells (eg. cancer cell viability) and each action represents a treatment strategy that the adaptive plasma system can take (eg. treatment duration, discharge voltage). MDP helps in defining the states, actions, and transition probabilities in the treatment process, but it does not inherently provide the optimal policy for selecting actions that lead to the best long-term rewards. Q-learning, a specific algorithm designed for reinforcement learning tasks [121], helps the adaptive plasma system to learn from its interactions with the environment and update its decision-making process by estimating the expected cumulative rewards for different actions in different states. This enables the system to make better decisions and optimize the treatment plan to achieve safer and more personalized cancer treatment (Figure 2). The study uses in vitro data to model cancer cell response, potentially limiting real-world applicability, but Gaussian processes aid in generalization. While the cancer treatment model as a Markov Decision Process lacks in-depth exploration of its limitations, it offers structured optimization with effective Q-learning adaptability. Safe Q-learning focuses on avoiding risky actions, leaving a gap in addressing other safety aspects but still enhances treatment safety. To enhance the model, future research should incorporate in vivo data, explore hybrid models, consider non-Markovian dynamics, investigate multi-agent systems, expand safety considerations, integrate real-time diagnostics, adopt personalized medicine, and validate through clinical trials for improved clinical relevance.
3.2. Gaussian process regression
Lin et al. [122] used Model Predictive Learning Control (MPLC), an extension of MPC that incorporates machine learning techniques for self-adaptive CAP jet optimization. While MPC typically uses a fixed mathematical model and optimization algorithm-based control strategy [67], MPLC integrates machine learning algorithms to capture system dynamics and adapt the control strategy based on real-time data. In the study, MPLC is implemented by using Gaussian Process (GP) regression, a statistical machine learning technique, to model the dynamic behavior of cancer cell viability in response to CAP treatment. The GP model is continuously updated with real-time feedback from the actual cancer cell responses, measured using real-time electrochemical impedance spectroscopy (EIS), after each treatment. This real-time feedback enables the MPLC algorithm to adapt and improve its predictions and treatment plans over time (Figure 3).
By dynamically adjusting the plasma treatment parameters based on actual system responses, the self-adaptive plasma jet can optimize its treatment strategy for a wide range of biomedical applications and variations in the system.
While the study demonstrates its use for cancer cell viability in response to CAP treatment, the transition to clinical applications may require additional research and validation. Pros include the adaptability of MPLC with GP regression, which enhances precision and effectiveness through real-time learning and feedback via electrochemical impedance spectroscopy (EIS). However, potential cons involve computational complexity and the reliability of GP models. Future directions should focus on clinical validation, scalability, and addressing computational challenges for practical healthcare adoption.
3.3. Deep learning
Deep learning uses artificial neural networks (ANNs) with multiple layers (deep neural networks) to make predictions from the labeled data (for instance real-time spectra from Optical Emission Spectroscopy). The self-adaptive plasma system empowered with AI holds great promise for advancing precision medicine and improving cancer treatment outcomes [119]. The self-adaptive plasma system is a plasma control system that can dynamically and autonomously diagnose plasma chemistry, adjust its operating parameters (eg. Gas and power inputs) to control and optimize plasma chemical composition in real-time according to the required species/dosage to kill cancer cells. This system typically employs artificial neural networks and Gradual-Mutation algorithms (GMA), to continuously monitor, analyze the datasets, control, and optimize plasma chemistry and thus make real-time decisions on how to modify the plasma conditions. The real-time spectra of plasma chemical composition and temperatures are obtained using spontaneous emission spectroscopy and processed by a diagnostic ANN. The diagnostic ANN provides valuable outputs on plasma chemistry, which are then used by a control ANN as inputs to adjust plasma parameters like gas injections and energy levels. The control ANN is trained by GMA to adjust the production of specific RONS associated with apoptosis of cancer cells. It receives feedback from the diagnostic ANN and dynamically adjusts the gas and power inputs to optimize the concentration of these species using in the plasma. GMA is used to train both the diagnostic and control ANNs effectively, allowing the self-adaptive plasma system to diagnose, control and optimize the plasma chemistry in real-time based on the OES data and specific objectives. Such an AI empowered self-adaptive plasma system allows for personalized treatments tailored to the specific characteristics of the patient's cancer
The integration of diagnostic ANN, chemical process network (CPN), and control ANN creates an adaptive and intelligent system for effective and personalized cancer treatment [123] (Figure 4).
AI-driven gas plasma can continuously learn from treatment outcomes, adapt to changing conditions, and optimize treatment plans. This personalized and dynamic approach has the potential to improve the efficacy of cancer therapies, reduce side effects, and advance precision medicine.
The absence of performance metrics and considerations is a critical limitation in the above discussed AI-based approaches. Performance evaluation is a fundamental aspect of any AI system. However, these research efforts primarily focus on the theoretical and conceptual aspects of AI-empowered plasma systems and their potential applications in cancer treatment. Given their early stage and limited access to real-world data, providing precise performance metrics is challenging. Addressing this gap necessitates future studies with interdisciplinary collaborations involving experts in computational science, machine learning, and medical engineering. These collaborations can facilitate the evaluation of computational efficiency, scalability, adaptability, and real-time processing capabilities of AI-empowered plasma systems. Additionally, benchmarking AI models against existing standards in the field enables a comparative analysis of efficiency and effectiveness. Adherence to established reporting standards and guidelines for AI research is essential for enhancing the rigor and impact of future studies.
As artificial intelligence gains broader acceptance in clinical applications, we foresee the imminent emergence of an artificial intelligence-driven adaptive plasma system for personalized cancer treatment, revolutionizing medical practice and paving the way for transformative advancements in oncology.
4. AI in Real-Time diagnostics
To exploit the adaptive and self-adaptive properties of CAP in developing personalized treatment protocols, it is important to estimate the operational parameters of the CAP sources and measure the cell response to CAP treatment in real-time.
4.1. Real-time diagnosis of operational parameters of CAP sources
Traditional diagnostics (eg. Laser induced fluorescence, mass spectrometry, spontaneous Raman scattering) of the different operational parameters of CAP sources are complex and expensive requiring sophisticated instrumentation setups and specialized analysis making them impractical for real-time use [124,125] and makes it challenging to control the plasma consistently and effectively. Using information-rich datasets collected from simpler diagnostic methods like optical emission spectroscopy (OES) [126], electro-acoustic emission and enabling fast, automated data processing and analysis using AI techniques in real-time monitoring and control, improves the feedback control, ensuring effective and optimal performance of CAP sources [124,125]. Simulated data [127,128], although not capturing all the complexities in real-world scenarios, complements real data by providing a controlled environment for analysis and experimentation.
Analysis of the datasets manually with traditional methods can be challenging due to complex relations and patterns in the data. ML and other computational approaches (Table 1) are designed to handle such complexity and can identify patterns and trends that may not be apparent to human analysis [125].
ML algorithms can be trained on the operational parameters and their corresponding data, allowing them to learn and generalize their knowledge for making predictions on new, unseen data. As new data is received the algorithms rapidly process and analyze data, enabling quick estimation of critical operational parameters from the spectral data or other input data in real-time. ML provides a promising pathway through the creation of input-output models that support real-time computations, thus enabling the implementation of more advanced feedback control strategies [117,135]. Constructing multivariable nonlinear prediction models to characterize the complex interactions between CAP and cancer cells is crucial for guiding effective cancer treatment approaches [133]. AI models can thus be used for real-time monitoring and diagnostics [124,129-131], detecting any deviations from the normal behavior [132,135], and predicting when maintenance or adjustment is required [119] for maintaining optimal real-time control of the operational parameters. Thus, AI- based approaches, specifically ML, can significantly improve the reliability and performance of CAP by adapting to dynamic and uncertain environments.
4.2. Real-time diagnosis of the cell responses to CAP treatment
Collecting real-time data on cancer cell responses to different CAP treatment modalities is challenging. Studies discussed in section 3.1 and 3.2 have utilized real-time EIS measurements [122,116] (Table 2) and has focused on incorporating automated feedback control systems into adaptive plasma system to optimize performance and safety (Figure 5).
Yet, advanced real-time diagnostic systems are still needed to provide sensitive, accurate, and reproducible data for these feedback control systems. To achieve optimal results, sophisticated diagnostic systems with high sensitivity, accuracy, and reproducibility are imperative. These systems should seamlessly integrate with the biological target and have minimal interference with the plasma treatment process. By fulfilling these criteria, the feedback control loop can make well-informed and precise adjustments, optimizing treatment outcomes for diverse cancer cells and individual patients. Developing electronic and optical sensors suitable for addressing this technological gap and to integrate these sensors into autonomous plasma systems must be focused [137]. Bridging this gap could lead to the development of next-generation medical plasma technologies, offering superior healthcare outcomes.
The applications of ML in modeling and simulating different aspects of CAP processing are well reviewed by Treischmann et al. [138]. Future advancements for CAP modelling and simulation promises precise control, pattern discovery, and transformative insights, advancing plasma science and technology. Open data access and collaboration are pivotal for breakthroughs [138].
5. Conclusions
Cancer is a challenging disease to treat due to its complexity, heterogeneity, invasion (metastasis), and resistance to treatments. The intricate nature of cancer manifests in the variability of responses to therapies among patients owing to the individualized genetic makeup and biological characteristics of their tumors. This necessitates tailored treatment approaches to optimize the chances of successful outcomes [139]. To achieve personalized treatment, with CAP is emerging as a promising approach as the parameters of the plasma device can be fine-tuned during the treatment to suit the patient’s specific dosage. CAP has shown to reprogram the tumor microenvironment [140-142] inhibiting tumor growth with high selectivity and even enhancing the effectiveness of other treatment modalities like chemotherapy [143-146], immunotherapy [147,148], radiotherapy [149-152], and nanomedicine [153-156] with improved treatment outcomes and minimized side-effects [157]. The self-adaptive nature of the adaptive plasma system complements the power of AI. As CAP devices can dynamically adjust treatment parameters during the therapy session, they can respond to real-time changes in tumor characteristics or patient physiology. AI-driven feedback loops can enhance this self-adaptation, ensuring that CAP treatment remains optimized and effective throughout the course of therapy, making it a promising avenue for achieving optimal treatment outcomes in personalized cancer care.
Better cross-disciplinary collaboration with plasma physicists, mathematicians, AI engineers, data scientist, oncologists, and clinicians can make these effective proposals achieve in the future that can be tailored towards personalized cancer treatment strategy. Advancing diagnostic tools, exploring new combination treatment strategies with CAP treatment, and modeling with AI will pave the way for more precise and personalized plasma-based therapies. AI is transforming plasma medicine, improving treatments with precise models, data analysis, real-time diagnostics, and protocols, despite ethical and transparency concerns [82]. The ongoing advancements at the frontiers of plasma technology in oncology are poised to revolutionize cancer treatment paradigms, providing new hope and improved outcomes for cancer patients worldwide. With continued efforts and interdisciplinary cooperation, plasma technology holds the potential to become an indispensable asset in the fight against cancer.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Martines, E. Plasma Technology for Biomedical Applications; MDPI, 2020.
- Duarte, S.; Panariello, B. H. D. Comprehensive Biomedical Applications of Low Temperature Plasmas. Arch. Biochem. Biophys. 2020, 693, 108560. [Google Scholar] [CrossRef]
- Bran\`y, D.; Dvorská, D.; Halašová, E.; Škovierová, H. Cold Atmospheric Plasma: A Powerful Tool for Modern Medicine. Int. J. Mol. Sci. 2020, 21, 2932. [Google Scholar] [CrossRef]
- Domonkos, M.; Tichá, P.; Trejbal, J.; Demo, P. Applications of Cold Atmospheric Pressure Plasma Technology in Medicine, Agriculture and Food Industry. Appl. Sci. 2021, 11, 4809. [Google Scholar] [CrossRef]
- Boehm, D.; Canal, C. Application of Plasma Technology in Bioscience and Biomedicine. Applied Sciences. MDPI 2021, p 7203.
- Laroussi, M.; Bekeschus, S.; Keidar, M.; Bogaerts, A.; Fridman, A.; Lu, X.; Ostrikov, K.; Hori, M.; Stapelmann, K.; Miller, V.; Reuter, S.; Laux, C.; Mesbah, A.; Walsh, J.; Jiang, C.; Thagard, S. M.; Tanaka, H.; Liu, D.; Yan, D.; Yusupov, M. Low-Temperature Plasma for Biology, Hygiene, and Medicine: Perspective and Roadmap. IEEE Trans. Radiat. Plasma Med. Sci. 2022, 6, 127–157. [Google Scholar] [CrossRef]
- Mumtaz, S.; Khan, R.; Rana, J. N.; Javed, R.; Iqbal, M.; Choi, E. H.; Han, I. Review on the Biomedical and Environmental Applications of Nonthermal Plasma. Catalysts 2023, 13, 685. [Google Scholar] [CrossRef]
- Bhattacharjee, B.; Bezbaruah, R.; Rynjah, D.; Newar, A.; Sengupta, S.; Pegu, P.; Dey, N.; Bora, S. C.; Barman, D. Cold Atmospheric Plasma: A Noteworthy Approach in Medical Science. Sci. Pharm. 2023, 2, 46–76. [Google Scholar] [CrossRef]
- Moszczyńska, J.; Roszek, K.; Wiśniewski, M. Non-Thermal Plasma Application in Medicine—Focus on Reactive Species Involvement. Int. J. Mol. Sci. 2023, 24, 12667. [Google Scholar] [CrossRef]
- Yan, D.; Wang, Q.; Adhikari, M.; Malyavko, A.; Lin, L.; Zolotukhin, D. B.; Yao, X.; Kirschner, M.; Sherman, J. H.; Keidar, M. A Physically Triggered Cell Death via Transbarrier Cold Atmospheric Plasma Cancer Treatment. ACS Appl. Mater. Interfaces 2020, 12, 34548–34563. [Google Scholar] [CrossRef]
- Tornin, J.; Labay, C.; Tampieri, F.; Ginebra, M.-P.; Canal, C. Evaluation of the Effects of Cold Atmospheric Plasma and Plasma-Treated Liquids in Cancer Cell Cultures. Nat. Protoc. 2021, 16, 2826–2850. [Google Scholar] [CrossRef]
- Suenaga, Y.; Takamatsu, T.; Aizawa, T.; Moriya, S.; Matsumura, Y.; Iwasawa, A.; Okino, A. Influence of Controlling Plasma Gas Species and Temperature on Reactive Species and Bactericidal Effect of the Plasma. Appl. Sci. 2021, 11(24), 11674. [Google Scholar] [CrossRef]
- Feibel, D.; Golda, J.; Held, J.; Awakowicz, P.; Schulz-von der Gathen, V.; Suschek, C. V.; Opländer, C.; Jansen, F. Gas Flow-Dependent Modification of Plasma Chemistry in ΜAPP Jet-Generated Cold Atmospheric Plasma and Its Impact on Human Skin Fibroblasts. Biomed. 2023, 11, 1242. [Google Scholar] [CrossRef]
- Lin, L.; Keidar, M. A Map of Control for Cold Atmospheric Plasma Jets: From Physical Mechanisms to Optimizations. Appl. Phys. Rev. 2021, 8. [Google Scholar] [CrossRef]
- Trelles, J. P. Pattern Formation and Self-Organization in Plasmas Interacting with Surfaces. J. Phys. D. Appl. Phys. 2016, 49, 393002. [Google Scholar] [CrossRef]
- Keidar,M. Adaptive and Self-Adaptive Plasma Cancer Therapeutic Platform. US patent, 11517366, 2022.
- Yan, D.; Cui, H.; Zhu, W.; Talbot, A.; Zhang, L. G.; Sherman, J. H.; Keidar, M. The Strong Cell-Based Hydrogen Peroxide Generation Triggered by Cold Atmospheric Plasma. Sci. Rep. 2017, 7, 10831. [Google Scholar] [CrossRef]
- Gjika, E.; Pal-Ghosh, S.; Tang, A.; Kirschner, M.; Tadvalkar, G.; Canady, J.; Stepp, M. A.; Keidar, M. Adaptation of Operational Parameters of Cold Atmospheric Plasma for in vitro Treatment of Cancer Cells. ACS Appl. Mater. Interfaces 2018, 10, 9269–9279. [Google Scholar] [CrossRef]
- Schweigert, I.; Zakrevsky, D.; Gugin, P.; Yelak, E.; Golubitskaya, E.; Troitskaya, O.; Koval, O. Interaction of Cold Atmospheric Argon and Helium Plasma Jets with Bio-Target with Grounded Substrate Beneath. Appl. Sci. 2019, 9, 4528. [Google Scholar] [CrossRef]
- Martinez, L.; Dhruv, A.; Balaras, E.; Keidar, M.; Martinez, L.; Dhruv, A.; Balaras, E.; Keidar, M. On Self Organization: Model for Ionization Wave Propagation with Targets of Varying Electrical Properties. PSST 2022, 31, 035004. [Google Scholar] [CrossRef]
- Keidar, M. Plasma for Cancer Treatment. Plasma Sources Sci. Technol. 2015, 24, 033001. [Google Scholar] [CrossRef]
- Keidar, M. Plasma Cancer Therapy, 1st ed.; Keidar, M., Ed.; Springer: Cham, 2020. [Google Scholar] [CrossRef]
- Alzahrani, E.; El-Dessoky, M. M.; Khan, M. A. Mathematical Model to Understand the Dynamics of Cancer, Prevention Diagnosis and Therapy. Mathematics 2023, 11, 1975. [Google Scholar] [CrossRef]
- Oke, S. I.; Matadi, M. B.; Xulu, S. S. Optimal Control Analysis of a Mathematical Model for Breast Cancer. Math. Comput. Appl. 2018, 23, 21. [Google Scholar]
- Shen, J.; Li, L.; Yang, T.; Cohen, P. S.; Sun, G. Biphasic Mathematical Model of Cell--Drug Interaction That Separates Target-Specific and off-Target Inhibition and Suggests Potent Targeted Drug Combinations for Multi-Driver Colorectal Cancer Cells. Cancers (Basel). 2020, 12, 436. [Google Scholar] [CrossRef]
- Hormuth, D. A.; Jarrett, A. M.; Davis, T.; Yankeelov, T. E. Towards an Image-Informed Mathematical Model of in Vivo Response to Fractionated Radiation Therapy. Cancers (Basel). 2021, 13, 1765. [Google Scholar] [CrossRef]
- Beck, R. J.; Weigelin, B.; Beltman, J. B. Mathematical Modelling Based on in Vivo Imaging Suggests CD137-Stimulated Cytotoxic T Lymphocytes Exert Superior Tumour Control Due to an Enhanced Antimitotic Effect on Tumour Cells. Cancers (Basel). 2021, 13, 2567. [Google Scholar] [CrossRef]
- Valle, P. A.; Coria, L. N.; Plata, C. Personalized Immunotherapy Treatment Strategies for a Dynamical System of Chronic Myelogenous Leukemia. Cancers (Basel). 2021, 13, 2030. [Google Scholar] [CrossRef]
- Anaya, D. A.; Dogra, P.; Wang, Z.; Haider, M.; Ehab, J.; Jeong, D. K.; Ghayouri, M.; Lauwers, G. Y.; Thomas, K.; Kim, R. ; others. A Mathematical Model to Estimate Chemotherapy Concentration at the Tumor-Site and Predict Therapy Response in Colorectal Cancer Patients with Liver Metastases. Cancers (Basel). 2021, 13, 444. [Google Scholar]
- Yonekura, Y.; Toki, H.; Watabe, T.; Kaneda-Nakashima, K.; Shirakami, Y.; Ooe, K.; Toyoshima, A.; Nakajima, H.; Tomiyama, N.; Bando, M. Mathematical Model for Evaluation of Tumor Response in Targeted Radionuclide Therapy with 211At Using Implanted Mouse Tumor. Int. J. Mol. Sci. 2022, 23, 15966. [Google Scholar] [CrossRef]
- Ghaffari Laleh, N.; Loeffler, C. M. L.; Grajek, J.; Sta\vnková, K.; Pearson, A. T.; Muti, H. S.; Trautwein, C.; Enderling, H.; Poleszczuk, J.; Kather, J. N. Classical Mathematical Models for Prediction of Response to Chemotherapy and Immunotherapy. PLoS Comput. Biol. 2022, 18, e1009822. [Google Scholar] [CrossRef]
- Italia, M.; Wertheim, K. Y.; Taschner-Mandl, S.; Walker, D.; Dercole, F. Mathematical Model of Clonal Evolution Proposes a Personalised Multi-Modal Therapy for High-Risk Neuroblastoma. Cancers (Basel). 2023, 15, 1986. [Google Scholar] [CrossRef]
- Jarrett, A. M.; Bloom, M. J.; Godfrey, W.; Syed, A. K.; Ekrut, D. A.; Ehrlich, L. I.; Yankeelov, T. E.; Sorace, A. G. Mathematical Modelling of Trastuzumab-Induced Immune Response in an in Vivo Murine Model of HER2+ Breast Cancer. Math. Med. Biol. a J. IMA 2019, 36, 381–410. [Google Scholar] [CrossRef]
- Budithi, A.; Su, S.; Kirshtein, A.; Shahriyari, L. Data Driven Mathematical Model of FOLFIRI Treatment for Colon Cancer. Cancers (Basel). 2021, 13, 2632. [Google Scholar] [CrossRef]
- Mohammad Mirzaei, N.; Su, S.; Sofia, D.; Hegarty, M.; Abdel-Rahman, M. H.; Asadpoure, A.; Cebulla, C. M.; Chang, Y. H.; Hao, W.; Jackson, P. R. ; others. A Mathematical Model of Breast Tumor Progression Based on Immune Infiltration. J. Pers. Med. 2021, 11, 1031. [Google Scholar]
- Bekker, R. A.; Kim, S.; Pilon-Thomas, S.; Enderling, H. Mathematical Modeling of Radiotherapy and Its Impact on Tumor Interactions with the Immune System. Neoplasia 2022, 28, 100796. [Google Scholar] [CrossRef] [PubMed]
- Bitsouni, V.; Tsilidis, V. Mathematical Modeling of Tumor-Immune System Interactions: The Effect of Rituximab on Breast Cancer Immune Response. J. Theor. Biol. 2022, 539, 111001. [Google Scholar] [CrossRef]
- Song, G.; Liang, G.; Tian, T.; Zhang, X. Mathematical Modeling and Analysis of Tumor Chemotherapy. Symmetry (Basel). 2022, 14, 704. [Google Scholar] [CrossRef]
- Jawad, S.; Winter, M.; Rahman, Z.-A. S. A.; Al-Yasir, Y. I. A.; Zeb, A. Dynamical Behavior of a Cancer Growth Model with Chemotherapy and Boosting of the Immune System. Mathematics 2023, 11, 406. [Google Scholar] [CrossRef]
- López-Alvarenga, J. C.; Minzoni-Alessio, A.; Olvera-Chávez, A.; Cruz-Pacheco, G.; Chimal-Eguia, J. C.; Hernández-Ru\’\iz, J.; Álvarez-Blanco, M. A.; Bautista-Hernández, M. Y.; Quispe-Siccha, R. M. A Mathematical Model to Optimize the Neoadjuvant Chemotherapy Treatment Sequence for Triple-Negative Locally Advanced Breast Cancer. Mathematics 2023, 11, 2410. [Google Scholar] [CrossRef]
- León-Triana, O.; Pérez-Mart\’\inez, A.; Ram\’\irez-Orellana, M.; Pérez-Garc\’\ia, V. M. Dual-Target CAR-Ts with on-and off-Tumour Activity May Override Immune Suppression in Solid Cancers: A Mathematical Proof of Concept. Cancers (Basel). 2021, 13, 703. [Google Scholar] [CrossRef]
- Sun, X.; Bao, J.; Shao, Y. Mathematical Modeling of Therapy-Induced Cancer Drug Resistance: Connecting Cancer Mechanisms to Population Survival Rates. Sci. Rep. 2016, 6, 22498. [Google Scholar] [CrossRef]
- Adhikarla, V.; Awuah, D.; Brummer, A. B.; Caserta, E.; Krishnan, A.; Pichiorri, F.; Minnix, M.; Shively, J. E.; Wong, J. Y. C.; Wang, X. ; others. A Mathematical Modeling Approach for Targeted Radionuclide and Chimeric Antigen Receptor t Cell Combination Therapy. Cancers (Basel). 2021, 13, 5171. [Google Scholar]
- Guzev, E.; Jadhav, S. S.; Hezkiy, E. E.; Sherman, M. Y.; Firer, M. A.; Bunimovich-Mendrazitsky, S. Validation of a Mathematical Model Describing the Dynamics of Chemotherapy for Chronic Lymphocytic Leukemia In Vivo. Cells 2022, 11, 2325. [Google Scholar] [CrossRef]
- Nave, Op.; Sigron, M. A Mathematical Model for the Treatment of Melanoma with the BRAF/MEK Inhibitor and Anti-PD-1. Appl. Sci. 2022, 12, 12474. [Google Scholar] [CrossRef]
- Nave, O. A Mathematical Model for Treatment Using Chemo-Immunotherapy. Heliyon 2022, 8. [Google Scholar] [CrossRef] [PubMed]
- Bozkurt, F.; Yousef, A.; Bilgil, H.; Baleanu, D. A Mathematical Model with Piecewise Constant Arguments of Colorectal Cancer with Chemo-Immunotherapy. Chaos, Solitons \& Fractals 2023, 168, 113207. [Google Scholar]
- Salim, S. S.; Malinzi, J.; Mureithi, E.; Shaban, N. Mathematical Modelling of Chemovirotherapy Cancer Treatment. Int. J. Model. Simul. 2023, 1–22. [Google Scholar] [CrossRef]
- Kim, Y.; Choe, B. Y.; Suh, T. S.; Sung, W. A Mathematical Model for Predicting Patient Responses to Combined Radiotherapy with CTLA-4 Immune Checkpoint Inhibitors. Cells 2023, 12. [Google Scholar] [CrossRef]
- Slavkova, K. P.; Patel, S. H.; Cacini, Z.; Kazerouni, A. S.; Gardner, A. L.; Yankeelov, T. E.; Hormuth, D. A. Mathematical Modelling of the Dynamics of Image-Informed Tumor Habitats in a Murine Model of Glioma. Sci. Rep. 2023, 13, 2916. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Thirumalai, D. A Mathematical Model for Phenotypic Heterogeneity in Breast Cancer with Implications for Therapeutic Strategies. J. R. Soc. Interface 2022, 19, 20210803. [Google Scholar] [CrossRef]
- Veith, T.; Schultz, A.; Alahmari, S.; Beck, R.; Johnson, J.; Andor, N. Mathematical Modeling of Clonal Interference by Density-Dependent Selection in Heterogeneous Cancer Cell Lines. Cells 2023, 12, 1849. [Google Scholar] [CrossRef]
- Khailov, E.; Grigorieva, E. Optimal Melanoma Treatment Protocols for a Bilinear Control Model. Mathematics 2023, 11, 3289. [Google Scholar] [CrossRef]
- Le, T.; Su, S.; Shahriyari, L. Investigating Optimal Chemotherapy Options for Osteosarcoma Patients through a Mathematical Model. Cells 2021, 10, 2009. [Google Scholar] [CrossRef]
- Phan, T.; Bennett, J.; Patten, T. Practical Understanding of Cancer Model Identifiability in Clinical Applications. Life 2023, 13, 410. [Google Scholar] [CrossRef]
- Vieira, L. C.; Costa, R. S.; Valério, D. An Overview of Mathematical Modelling in Cancer Research: Fractional Calculus as Modelling Tool. Fractal and Fractional. Multidisciplinary Digital Publishing Institute August 1, 2023, p 595. [CrossRef]
- Uçar, E.; Özdemir, N. New Fractional Cancer Mathematical Model via IL-10 Cytokine and Anti-PD-L1 Inhibitor. Fractal Fract. 2023, 7, 151. [Google Scholar] [CrossRef]
- Farman, M.; Batool, M.; Nisar, K. S.; Ghaffari, A. S.; Ahmad, A. Controllability and Analysis of Sustainable Approach for Cancer Treatment with Chemotherapy by Using the Fractional Operator. Results Phys. 2023, 106630. [Google Scholar] [CrossRef]
- Elharrar, X.; Barbolosi, D.; Ciccolini, J.; Meille, C.; Faivre, C.; Lacarelle, B.; André, N.; Barlesi, F. A Phase Ia/Ib Clinical Trial of Metronomic Chemotherapy Based on a Mathematical Model of Oral Vinorelbine in Metastatic Non-Small Cell Lung Cancer and Malignant Pleural Mesothelioma: Rationale and Study Protocol. BMC Cancer 2016, 1. [Google Scholar] [CrossRef] [PubMed]
- Smalley, I.; Kim, E.; Li, J.; Spence, P.; Wyatt, C. J.; Eroglu, Z.; Sondak, V. K.; Messina, J. L.; Babacan, N. A.; Maria-Engler, S. S.; De Armas, L.; Williams, S. L.; Gatenby, R. A.; Chen, Y. A.; Anderson, A. R. A.; Smalley, K. S. M. Leveraging Transcriptional Dynamics to Improve BRAF Inhibitor Responses in Melanoma. EBioMedicine 2019, 48, 178–190. [Google Scholar] [CrossRef]
- Guerreiro, N.; Jullion, A.; Ferretti, S.; Fabre, C.; Meille, C. Translational Modeling of Anticancer Efficacy to Predict Clinical Outcomes in a First-in-Human Phase 1 Study of MDM2 Inhibitor HDM201. AAPS J. 2021, 23(2), 1–17. [Google Scholar] [CrossRef]
- Brüningk, S. C.; Peacock, J.; Whelan, C. J.; Brady-Nicholls, R.; Yu, H. H. M.; Sahebjam, S.; Enderling, H. Intermittent Radiotherapy as Alternative Treatment for Recurrent High Grade Glioma: A Modeling Study Based on Longitudinal Tumor Measurements. Sci. Rep. 2021, 11. [Google Scholar] [CrossRef]
- Mathur, D.; Barnett, E.; Scher, H. I.; Xavier, J. B. Optimizing the Future: How Mathematical Models Inform Treatment Schedules for Cancer. Trends Cancer 2022, 8, 506. [Google Scholar] [CrossRef] [PubMed]
- Leder, K.; Pitter, K.; Laplant, Q.; Hambardzumyan, D.; Ross, B. D.; Chan, T. A.; Holland, E. C.; Michor, F. Mathematical Modeling of Pdgf-Driven Glioblastoma Reveals Optimized Radiation Dosing Schedules. Cell, 2014, 156, 603–616. [Google Scholar] [CrossRef]
- Dean, J. A.; Tanguturi, S. K.; Cagney, D.; Shin, K. Y.; Youssef, G.; Aizer, A.; Rahman, R.; Hammoudeh, L.; Reardon, D.; Lee, E.; Dietrich, J.; Tamura, K.; Aoyagi, M.; Wickersham, L.; Wen, P. Y.; Catalano, P.; Haas-Kogan, D.; Alexander, B. M.; Michor, F. Phase I Study of a Novel Glioblastoma Radiation Therapy Schedule Exploiting Cell-State Plasticity. Neuro. Oncol. 2023, 25, 1100–1112. [Google Scholar] [CrossRef]
- Keidar, M.; Yan, D.; Beilis, I. I.; Trink, B.; Sherman, J. H. Plasmas for Treating Cancer: Opportunities for Adaptive and Self-Adaptive Approaches. Trends Biotechnol. 2018, 36, 586–593. [Google Scholar] [CrossRef]
- Lyu, Y.; Lin, L.; Gjika, E.; Lee, T.; Keidar, M. Mathematical Modeling and Control for Cancer Treatment with Cold Atmospheric Plasma Jet. J. Phys. D. Appl. Phys. 2019, 51, 185202. [Google Scholar] [CrossRef]
- Tanaka, H.; Bekeschus, S.; Yan, D.; Hori, M. ; Keidar,M; Laroussi,M. Plasma-Treated Solutions (PTS) in Cancer Therapy. Cancers 2021, 13, 1737. [Google Scholar] [CrossRef] [PubMed]
- Tampieri, F.; Gorbanev, Y.; Sardella, E. Plasma-Treated Liquids in Medicine: Let’s Get Chemical. Plasma Process. Polym. 2023, e2300077. [Google Scholar] [CrossRef]
- Stache, A. B.; Mih\uail\ua, I.; Gerber, I. C.; Dragoș, L. M.; Mihai, C. T.; Ivanov, I. C.; Topal\ua, I.; Gorgan, D.-L. Optimization of Indirect CAP Exposure as an Effective Osteosarcoma Cells Treatment with Cytotoxic Effects. Appl. Sci. 2023, 13, 7803. [Google Scholar] [CrossRef]
- Solé-Martí, X.; Vilella, T.; Labay, C.; Tampieri, F.; Ginebra, M. P.; Canal, C. Thermosensitive Hydrogels to Deliver Reactive Species Generated by Cold Atmospheric Plasma: A Case Study with Methylcellulose. Biomater. Sci. 2022, 10, 3845–3855. [Google Scholar] [CrossRef]
- Malyavko, A.; Yan, D.; Wang, Q.; Klein, A.L.; Patel, K.C.; Sherman, J.H.; Keidar, M. Cold Atmospheric Plasma Cancer Treatment, Direct versus Indirect Approaches. Mater. Adv. 2020, 1, 1494–1505. [Google Scholar] [CrossRef]
- Poramapijitwat, P.; Thana, P.; Sukum, P.; Liangdeng, Y.; Kuensaen, C.; Boonyawan, D. Selective Cytotoxicity of Lung Cancer Cells—A549 and H1299—Induced by Ringer’s Lactate Solution Activated by a Non-Thermal Air Plasma Jet Device, Nightingale®. Plasma Chem. Plasma Process. 2023, 43, 805–830. [Google Scholar] [CrossRef]
- Miebach, L.; Mohamed, H.; Wende, K.; Miller, V.; Bekeschus, S. Pancreatic Cancer Cells Undergo Immunogenic Cell Death upon Exposure to Gas Plasma-Oxidized Ringers Lactate. Cancers 2023, 15, 319. [Google Scholar] [CrossRef]
- Pavlik, T.; Gudkova, V.; Razvolyaeva, D.; Pavlova, M.; Kostukova, N.; Miloykovich, L.; Kolik, L.; Konchekov, E.; Shimanovskii, N. The Role of Autophagy and Apoptosis in the Combined Action of Plasma-Treated Saline, Doxorubicin, and Medroxyprogesterone Acetate on K562 Myeloid Leukaemia Cells. Int. J. Mol. Sci. 2023, 24, 5100. [Google Scholar] [CrossRef]
- Wang, X.; Liu, N.; Liu, J.; Cui, Y.; Wang, X.; Lu, J.; Kang, C.; Gao, L.; Shi, X.; Zhang, G. Comparison of Direct and Indirect Low-Temperature Plasma Triggering Immunogenic Cell Death in B16F10 Melanoma. Plasma Process. Polym. 2023, 20, e2200206. [Google Scholar] [CrossRef]
- Bengtson, C.; Bogaerts, A. On the Anti-Cancer Effect of Cold Atmospheric Plasma and the Possible Role of Catalase-Dependent Apoptotic Pathways. Cells 2020, 9. [Google Scholar] [CrossRef]
- Bengtson, C.; Bogaerts, A. The Quest to Quantify Selective and Synergistic Effects of Plasma for Cancer Treatment: Insights from Mathematical Modeling. Int. J. Mol. Sci. 2021, 22, 5033. [Google Scholar] [CrossRef]
- Martemucci, G.; Costagliola, C.; Mariano, M.; D’andrea, L.; Napolitano, P.; D’Alessandro, A. G. Free Radical Properties, Source and Targets, Antioxidant Consumption and Health. Oxygen 2022, 2, 48–78. [Google Scholar] [CrossRef]
- Rabaan, A. A.; Bakhrebah, M. A.; AlSaihati, H.; Alhumaid, S.; Alsubki, R. A.; Turkistani, S. A.; Al-Abdulhadi, S.; Aldawood, Y.; Alsaleh, A. A.; Alhashem, Y. N. ; others. Artificial Intelligence for Clinical Diagnosis and Treatment of Prostate Cancer. Cancers (Basel). 2022, 14, 5595. [Google Scholar]
- Koteluk, O.; Wartecki, A.; Mazurek, S.; Kołodziejczak, I.; Mackiewicz, A. How Do Machines Learn? Artificial Intelligence as a New Era in Medicine. J. Pers. Med. 2021, 11, 32. [Google Scholar]
- Ercan, U. K.; Özdemir, G. D.; Özdemir, M. A.; Güren, O. Plasma Medicine: The Era of Artificial Intelligence. Plasma Process. Polym. 2023, e2300066. [Google Scholar] [CrossRef]
- Galuzio, P. P.; Cherif, A. Recent Advances and Future Perspectives in the Use of Machine Learning and Mathematical Models in Nephrology. Adv. Chronic Kidney Dis. 2022, 29, 472–479. [Google Scholar] [CrossRef]
- Zhang, B.; Shi, H.; Wang, H. Machine Learning and AI in Cancer Prognosis, Prediction, and Treatment Selection: A Critical Approach. J. Multidiscip. Healthc. 2023, 16, 1779. [Google Scholar] [CrossRef] [PubMed]
- Bonzanini, A. D.; Shao, K.; Graves, D. B.; Hamaguchi, S.; Mesbah, A. Foundations of Machine Learning for Low-Temperature Plasmas: Methods and Case Studies. Plasma Sources Sci. Technol. 2023, 32, 024003. [Google Scholar] [CrossRef]
- Kim, S.; Jung, S.; Park, Y.; Lee, J.; Park, J. Effective Liver Cancer Diagnosis Method Based on Machine Learning Algorithm. In 2014 7th International Conference on Biomedical Engineering and Informatics; 2014; pp 714–718.
- Glučina, M.; Lorencin, A.; An\djelić, N.; Lorencin, I. Cervical Cancer Diagnostics Using Machine Learning Algorithms and Class Balancing Techniques. Appl. Sci. 2023, 13, 1061. [Google Scholar] [CrossRef]
- Gawade, S.; Bhansali, A.; Patil, K.; Shaikh, D. Application of the Convolutional Neural Networks and Supervised Deep-Learning Methods for Osteosarcoma Bone Cancer Detection. Healthc. Anal. 2023, 3, 100153. [Google Scholar] [CrossRef]
- Barata, C.; Rotemberg, V.; Codella, N. C. F.; Tschandl, P.; Rinner, C.; Akay, B. N.; Apalla, Z.; Argenziano, G.; Halpern, A.; Lallas, A.; et al. A Reinforcement Learning Model for AI-Based Decision Support in Skin Cancer. Nat. Med. 2023, 1–6. [Google Scholar] [CrossRef]
- Mengash, H. A.; Alamgeer, M.; Maashi, M.; Othman, M.; Hamza, M. A.; Ibrahim, S. S.; Zamani, A. S.; Yaseen, I. Leveraging Marine Predators Algorithm with Deep Learning for Lung and Colon Cancer Diagnosis. Cancers (Basel). 2023, 15, 1591. [Google Scholar] [CrossRef]
- Afrash, M. R.; Mirbagheri, E.; Mashoufi, M.; Kazemi-Arpanahi, H. Optimizing Prognostic Factors of Five-Year Survival in Gastric Cancer Patients Using Feature Selection Techniques with Machine Learning Algorithms: A Comparative Study. BMC Med. Inform. Decis. Mak. 2023, 23, 54. [Google Scholar] [CrossRef] [PubMed]
- Pais, R. J.; Lopes, F.; Parreira, I.; Silva, M.; Silva, M.; Moutinho, M. G. Predicting Cancer Prognostics from Tumour Transcriptomics Using an Auto Machine Learning Approach. In Medical Sciences Forum; 2023; Vol. 22, p 6.
- Bostanci, E.; Kocak, E.; Unal, M.; Guzel, M. S.; Acici, K.; Asuroglu, T. Machine Learning Analysis of RNA-Seq Data for Diagnostic and Prognostic Prediction of Colon Cancer. Sensors 2023, 23, 3080. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Ye, B.; Wu, L.; Ni, S.; Li, Y.; Wang, Q.; Zhang, P.; Wang, D. Machine Learning-Based Prediction of Survival Prognosis in Esophageal Squamous Cell Carcinoma. Sci. Rep. 2023, 13, 13532. [Google Scholar] [CrossRef]
- Wu, R.; Luo, J.; Wan, H.; Zhang, H.; Yuan, Y.; Hu, H.; Feng, J.; Wen, J.; Wang, Y.; Li, J. ; others. Evaluation of Machine Learning Algorithms for the Prognosis of Breast Cancer from the Surveillance, Epidemiology, and End Results Database. PLoS One 2023, 18, e0280340. [Google Scholar] [CrossRef]
- Botlagunta, M.; Botlagunta, M. D.; Myneni, M. B.; Lakshmi, D.; Nayyar, A.; Gullapalli, J. S.; Shah, M. A. Classification and Diagnostic Prediction of Breast Cancer Metastasis on Clinical Data Using Machine Learning Algorithms. Sci. Rep. 2023, 13, 485. [Google Scholar] [CrossRef]
- Tian, H.; Liu, Z.; Liu, J.; Zong, Z.; Chen, Y.; Zhang, Z.; Li, H. Application of Machine Learning Algorithm in Predicting Distant Metastasis of T1 Gastric Cancer. Sci. Rep. 2023, 13, 5741. [Google Scholar] [CrossRef]
- Mengash, H. A.; Alamgeer, M.; Maashi, M.; Othman, M.; Hamza, M. A.; Ibrahim, S. S.; Zamani, A. S.; Yaseen, I. Leveraging Marine Predators Algorithm with Deep Learning for Lung and Colon Cancer Diagnosis. Cancers (Basel). 2023, 15, 1591. [Google Scholar] [CrossRef]
- Prelaj, A.; Boeri, M.; Robuschi, A.; Ferrara, R.; Proto, C.; Lo Russo, G.; Galli, G.; De Toma, A.; Brambilla, M.; Occhipinti, M. ; others. Machine Learning Using Real-World and Translational Data to Improve Treatment Selection for NSCLC Patients Treated with Immunotherapy. Cancers (Basel). 2022, 14, 435. [Google Scholar]
- Kong, J.; Lee, H.; Kim, D.; Han, S. K.; Ha, D.; Shin, K.; Kim, S. Network-Based Machine Learning in Colorectal and Bladder Organoid Models Predicts Anti-Cancer Drug Efficacy in Patients. Nat. Commun. 2020, 11, 5485. [Google Scholar] [CrossRef]
- Chen, S.; Shu, Z.; Li, Y.; Chen, B.; Tang, L.; Mo, W.; Shao, G.; Shao, F. Machine Learning-Based Radiomics Nomogram Using Magnetic Resonance Images for Prediction of Neoadjuvant Chemotherapy Efficacy in Breast Cancer Patients. Front. Oncol. 2020, 10, 1410. [Google Scholar] [CrossRef]
- Shao, Y.; Dang, Y.; Cheng, Y.; Gui, Y.; Chen, X.; Chen, T.; Zeng, Y.; Tan, L.; Zhang, J.; Xiao, M. ; others. Predicting the Efficacy of Neoadjuvant Chemotherapy for Pancreatic Cancer Using Deep Learning of Contrast-Enhanced Ultrasound Videos. Diagnostics 2023, 13, 2183. [Google Scholar]
- Arezzo, F.; La Forgia, D.; Venerito, V.; Moschetta, M.; Tagliafico, A. S.; Lombardi, C.; Loizzi, V.; Cicinelli, E.; Cormio, G. A Machine Learning Tool to Predict the Response to Neoadjuvant Chemotherapy in Patients with Locally Advanced Cervical Cancer. Appl. Sci. 2021, 11, 823. [Google Scholar] [CrossRef]
- Johannet, P.; Coudray, N.; Donnelly, D. M.; Jour, G.; Illa-Bochaca, I.; Xia, Y.; Johnson, D. B.; Wheless, L.; Patrinely, J. R.; Nomikou, S. ; others. Using Machine Learning Algorithms to Predict Immunotherapy Response in Patients with Advanced Melanoma. Clin. Cancer Res. 2021, 27, 131–140. [Google Scholar] [PubMed]
- Qureshi, R.; Basit, S. A.; Shamsi, J. A.; Fan, X.; Nawaz, M.; Yan, H.; Alam, T. Machine Learning Based Personalized Drug Response Prediction for Lung Cancer Patients. Sci. Rep. 2022, 12, 18935. [Google Scholar] [CrossRef]
- Kong, J.; Ha, D.; Lee, J.; Kim, I.; Park, M.; Im, S.-H.; Shin, K.; Kim, S. Network-Based Machine Learning Approach to Predict Immunotherapy Response in Cancer Patients. Nat. Commun. 2022, 13, 3703. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Bo, Z.; Zhao, Z.; Yang, J.; Yang, Y.; Li, H.; Yang, Y.; Wang, J.; Su, Q.; Wang, J. ; others. Machine Learning to Predict the Response to Lenvatinib Combined with Transarterial Chemoembolization for Unresectable Hepatocellular Carcinoma. Cancers (Basel). 2023, 15, 625. [Google Scholar]
- Fujima, N.; Shimizu, Y.; Yoshida, D.; Kano, S.; Mizumachi, T.; Homma, A.; Yasuda, K.; Onimaru, R.; Sakai, O.; Kudo, K. ; others. Machine-Learning-Based Prediction of Treatment Outcomes Using MR Imaging-Derived Quantitative Tumor Information in Patients with Sinonasal Squamous Cell Carcinomas: A Preliminary Study. Cancers (Basel). 2019, 11, 800. [Google Scholar]
- Arezzo, F.; La Forgia, D.; Venerito, V.; Moschetta, M.; Tagliafico, A. S.; Lombardi, C.; Loizzi, V.; Cicinelli, E.; Cormio, G. A Machine Learning Tool to Predict the Response to Neoadjuvant Chemotherapy in Patients with Locally Advanced Cervical Cancer. Appl. Sci. 2021, 11, 823. [Google Scholar] [CrossRef]
- Abuhelwa, A. Y.; Kichenadasse, G.; McKinnon, R. A.; Rowland, A.; Hopkins, A. M.; Sorich, M. J. Machine Learning for Prediction of Survival Outcomes with Immune-Checkpoint Inhibitors in Urothelial Cancer. Cancers (Basel). 2021, 13, 2001. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Brendel, M.; Wu, N.; Ge, W.; Zhang, H.; Rietschel, P.; Quek, R. G. W.; Pouliot, J.-F.; Wang, F.; Harnett, J. Machine Learning Models for Identifying Predictors of Clinical Outcomes with First-Line Immune Checkpoint Inhibitor Therapy in Advanced Non-Small Cell Lung Cancer. Sci. Rep. 2022, 12, 17670. [Google Scholar] [CrossRef]
- Qu, J.; Li, C.; Liu, M.; Wang, Y.; Feng, Z.; Li, J.; Wang, W.; Wu, F.; Zhang, S.; Zhao, X. Prognostic Models Using Machine Learning Algorithms and Treatment Outcomes of Occult Breast Cancer Patients. J. Clin. Med. 2023, 12, 3097. [Google Scholar] [CrossRef] [PubMed]
- Savić, M.; Kurbalija, V.; Ilić, M.; Ivanović, M.; Jakovetić, D.; Valachis, A.; Autexier, S.; Rust, J.; Kosmidis, T. The Application of Machine Learning Techniques in Prediction of Quality-of-Life Features for Cancer Patients. Comput. Sci. Inf. Syst. 2023, 20, 381–404. [Google Scholar] [CrossRef]
- Liao, J.; Li, X.; Gan, Y.; Han, S.; Rong, P.; Wang, W.; Li, W.; Zhou, L. Artificial Intelligence Assists Precision Medicine in Cancer Treatment. Front. Oncol. 2023, 12, 998222. [Google Scholar] [CrossRef] [PubMed]
- Charalambous, A.; Dodlek, N. Big Data, Machine Learning, and Artificial Intelligence to Advance Cancer Care: Opportunities and Challenges. Semin. Oncol. Nurs. 2023, 39. [Google Scholar] [CrossRef]
- Hou, Z.; Lee, T.; Keidar, M. Reinforcement Learning with Safe Exploration for Adaptive Plasma Cancer Treatment. IEEE Trans. Radiat. Plasma Med. Sci. 2022, 6, 482–492. [Google Scholar] [CrossRef]
- Bonzanini, A. D.; Shao, K.; Stancampiano, A.; Graves, D. B.; Mesbah, A. Perspectives on Machine Learning-Assisted Plasma Medicine: Toward Automated Plasma Treatment. IEEE Trans. Radiat. Plasma Med. Sci. 2022, 6, 16–32. [Google Scholar] [CrossRef]
- Chan, K. J.; Makrygiorgos, G.; Mesbah, A. Towards Personalized Plasma Medicine via Data-Efficient Adaptation of Fast Deep Learning-Based MPC Policies. Am. Control Conf. 2023, 2769–2775. [Google Scholar] [CrossRef]
- Lin, L.; Yan, D.; Lee, T.; Keidar, M. Self-Adaptive Plasma Chemistry and Intelligent Plasma Medicine. Adv. Intell. Syst. 2022, 4, 2100112. [Google Scholar] [CrossRef]
- Littman, M. L.; Szepesvári, C. A Generalized Reinforcement-Learning Model: Convergence and Applications. In ICML, 1996, 310–318.
- Chen, C. L.; Dong, D. Y.; Li, H. X.; Tarn, T. J. Hybrid MDP Based Integrated Hierarchical Q-Learning. Sci. China Inf. Sci. 2011, 54, 2279–2294. [Google Scholar] [CrossRef]
- Lin, L.; Hou, Z.; Yao, X.; Liu, Y.; Sirigiri, J. R.; Lee, T.; Keidar, M. Introducing Adaptive Cold Atmospheric Plasma: The Perspective of Adaptive Cold Plasma Cancer Treatments Based on Real-Time Electrochemical Impedance Spectroscopy. Phys. Plasmas 2020, 27, 063501. [Google Scholar] [CrossRef]
- Lin, L.; Keidar, M. Artificial Intelligence without Digital Computers: Programming Matter at a Molecular Scale. Adv. Intell. Syst. 2022, 4, 2200157. [Google Scholar] [CrossRef]
- Gidon, D.; Pei, X.; Bonzanini, A. D.; Graves, D. B.; Mesbah, A. Machine Learning for Real-Time Diagnostics of Cold Atmospheric Plasma Sources. IEEE Trans. Radiat. Plasma Med. Sci. 2019, 3(5), 597–605. [Google Scholar] [CrossRef]
- Mesbah, A.; Graves, D. B. Machine Learning for Modeling, Diagnostics, and Control of Non-Equilibrium Plasmas. J. Phys. D. Appl. Phys. 2019, 52, 30LT02. [Google Scholar] [CrossRef]
- Zaplotnik, R.; Primc, G.; Vesel, A. Optical Emission Spectroscopy as a Diagnostic Tool for Characterization of Atmospheric Plasma Jets. Appl. Sci. 2021, 11, 2275. [Google Scholar] [CrossRef]
- Witman, M.; Gidon, D.; Graves, D. B.; Smit, B.; Mesbah, A. Sim-to-Real Transfer Reinforcement Learning for Control of Thermal Effects of an Atmospheric Pressure Plasma Jet. Plasma Sources Sci. Technol. 2019, 28. [Google Scholar] [CrossRef]
- Zhang, Y. T.; Gao, S. H.; Ai, F. Efficient Numerical Simulation of Atmospheric Pulsed Discharges by Introducing Deep Learning. Front. Phys. 2023, 11. [Google Scholar] [CrossRef]
- van Der Gaag, T.; Onishi, H.; Akatsuka, H. Arbitrary EEDF Determination of Atmospheric-Pressure Plasma by Applying Machine Learning to OES Measurement. Phys. Plasmas 2021, 28. [Google Scholar] [CrossRef]
- van Der Gaag, T.; Nezu, A.; Akatsuka, H. Practical Considerations of the Visible Bremsstrahlung Inversion (VBI) Method for Arbitrary EEDF Determination in Cold Atmospheric-Pressure Plasma. Jpn. J. Appl. Phys. 2022, 61. [Google Scholar] [CrossRef]
- van der Gaag, T.; Nezu, A.; Akatsuka, H. Partial EEDF Analysis and Electron Diagnostics of Atmospheric-Pressure Argon and Argon-Helium DBD Plasma. J. Phys. D. Appl. Phys. 2023, 56. [Google Scholar] [CrossRef]
- Chang, J.; Niu, P. H.; Chen, C. W.; Cheng, Y. C. Using Deep Convolutional Neural Networks to Classify the Discharge Current of a Cold Atmospheric-Pressure Plasma Jet. IEEE Trans. Plasma Sci. 2023, 51, 311–319. [Google Scholar] [CrossRef]
- Lazarus, M.; Yan, D.; Limanowski, R.; Lin, L.; Keidar, M. Recognizing Cold Atmospheric Plasma Plume Using Computer Vision. Plasma 2022, 5, 341–350. [Google Scholar] [CrossRef]
- Lin, L.; Gershman, S.; Raitses, Y.; Keidar, M. Multi-Scale Plasma Chemistry Using Physics-Informed Neural Network. J. Phys D Appl. Phys.in review.
- Kim, D. H.; Hong, S. J. Use of Plasma Information in Machine-Learning-Based Fault Detection and Classification for Advanced Equipment Control. IEEE Trans. Semicond. Manuf. 2021, 34, 408–419. [Google Scholar] [CrossRef]
- Sebastian, A.; Lipa, D.; Ptasinska, S. DNA Strand Breaks and Denaturation as Probes of Chemical Reactivity versus Thermal Effects of Atmospheric Pressure Plasma Jets. ACS Omega 2022. [CrossRef]
- Sabrin, S.; Karmokar, D. K.; Karmakar, N. C.; Hong, S. H.; Habibullah, H.; Szili, E. J. Opportunities of Electronic and Optical Sensors in Autonomous Medical Plasma Technologies. ACS Sensors 2023, 8, 974–993. [Google Scholar] [CrossRef]
- Trieschmann, J.; Vialetto, L.; Gergs, T. Machine Learning for Advancing Low-Temperature Plasma Modeling and Simulation. 2023. [CrossRef]
- Dey, A.; Mitra, A.; Pathak, S.; Prasad, S.; Zhang, A. S.; Zhang, H.; Sun, X. F.; Banerjee, A. Recent Advancements, Limitations, and Future Perspectives of the Use of Personalized Medicine in Treatment of Colon Cancer. Technol. Cancer Res. Treat. 2023, 22. [Google Scholar] [CrossRef]
- Aggelopoulos, C. A.; Christodoulou, A.-M.; Tachliabouri, M.; Meropoulis, S.; Christopoulou, M.-E.; Karalis, T. T.; Chatzopoulos, A.; Skandalis, S. S. Cold Atmospheric Plasma Attenuates Breast Cancer Cell Growth through Regulation of Cell Microenvironment Effectors. Front. Oncol. 2022, 11, 826865. [Google Scholar] [CrossRef] [PubMed]
- Byun, J.; Wu, Y.; Lee, J.; Kim, J. S.; Shim, G.; Oh, Y.-K. External Cold Atmospheric Plasma-Responsive on-Site Hydrogel for Remodeling Tumor Immune Microenvironment. Biomaterials 2023, 299, 122162. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhu, K. Cold Atmospheric Plasma: Novel Opportunities for Tumor Microenvironment Targeting. Cancer Med. 2023, 12, 7189–7206. [Google Scholar] [CrossRef]
- Patrakova, E.; Biryukov, M.; Troitskaya, O.; Gugin, P.; Milakhina, E.; Semenov, D.; Poletaeva, J.; Ryabchikova, E.; Novak, D.; Kryachkova, N. ; others. Chloroquine Enhances Death in Lung Adenocarcinoma A549 Cells Exposed to Cold Atmospheric Plasma Jet. Cells 2023, 12, 290. [Google Scholar] [PubMed]
- Kniazeva, V.; Tzerkovsky, D.; Baysal, Ö.; Kornev, A.; Roslyakov, E.; Kostevitch, S. Adjuvant Composite Cold Atmospheric Plasma Therapy Increases Antitumoral Effect of Doxorubicin Hydrochloride. Front. Oncol. 2023, 13, 1171042. [Google Scholar] [CrossRef]
- Nitsch, A.; Qarqash, S.; Römer, S.; Schoon, J.; Ekkernkamp, A.; Niethard, M.; Reichert, J. C.; Wassilew, G. I.; Tzvetkov, M. V; Haralambiev, L. Enhancing the Impact of Chemotherapy on Ewing Sarcoma Cells through Combination with Cold Physical Plasma. Int. J. Mol. Sci. 2023, 24, 8669. [Google Scholar] [CrossRef] [PubMed]
- Soni, V.; Adhikari, M.; Lin, L.; Sherman, J. H.; Keidar, M. Theranostic Potential of Adaptive Cold Atmospheric Plasma with Temozolomide to Checkmate Glioblastoma: An In Vitro Study. Cancers (Basel). 2022, 14, 3116. [Google Scholar] [CrossRef] [PubMed]
- Fang, T.; Cao, X.; Shen, B.; Chen, Z.; Chen, G. Injectable Cold Atmospheric Plasma-Activated Immunotherapeutic Hydrogel for Enhanced Cancer Treatment. Biomaterials 2023, 122189. [Google Scholar] [CrossRef]
- Chen, G.; Chen, Z.; Wen, D.; Wang, Z.; Li, H.; Zeng, Y.; Dotti, G.; Wirz, R. E.; Gu, Z. Transdermal Cold Atmospheric Plasma-Mediated Immune Checkpoint Blockade Therapy. Proc. Natl. Acad. Sci. 2020, 117, 3687–3692. [Google Scholar] [CrossRef]
- Momeni, S.; Shanei, A.; Sazgarnia, A.; Attaran, N.; Aledavood, S. A. The Synergistic Effect of Cold Atmospheric Plasma Mediated Gold Nanoparticles Conjugated with Indocyanine Green as an Innovative Approach to Cooperation with Radiotherapy. Cell J. 2023, 25, 51. [Google Scholar]
- Kenari, A. J.; Siadati, S. N.; Abedian, Z.; Sohbatzadeh, F.; Amiri, M.; Gorji, K. E.; Babapour, H.; Zabihi, E.; Ghoreishi, S. M.; Mehraeen, R. ; others. Therapeutic Effect of Cold Atmospheric Plasma and Its Combination with Radiation as a Novel Approach on Inhibiting Cervical Cancer Cell Growth (HeLa Cells). Bioorg. Chem. 2021, 111, 104892. [Google Scholar]
- Pansare, K.; Vaid, A.; Singh, S. R.; Rane, R.; Visani, A.; Ranjan, M.; Krishna, C. M.; Sarin, R.; Joseph, A. Effect of Cold Atmospheric Plasma Jet and Gamma Radiation Treatments on Gingivobuccal Squamous Cell Carcinoma and Breast Adenocarcinoma Cells. Plasma Chem. Plasma Process. 2021, 1–16. [Google Scholar] [CrossRef]
- Pasqual-Melo, G.; Sagwal, S. K.; Freund, E.; Gandhirajan, R. K.; Frey, B.; von Woedtke, T.; Gaipl, U.; Bekeschus, S. Combination of Gas Plasma and Radiotherapy Has Immunostimulatory Potential and Additive Toxicity in Murine Melanoma Cells in Vitro. Int. J. Mol. Sci. 2020, 21, 1379. [Google Scholar] [CrossRef]
- He, Z.; Liu, K.; Manaloto, E.; Casey, A.; Cribaro, G. P.; Byrne, H. J.; Tian, F.; Barcia, C.; Conway, G. E.; Cullen, P. J. ; others. Cold Atmospheric Plasma Induces ATP-Dependent Endocytosis of Nanoparticles and Synergistic U373MG Cancer Cell Death. Sci. Rep. 2018, 8, 5298. [Google Scholar]
- Li, W.; Yu, H.; Ding, D.; Chen, Z.; Wang, Y.; Wang, S.; Li, X.; Keidar, M.; Zhang, W. Cold Atmospheric Plasma and Iron Oxide-Based Magnetic Nanoparticles for Synergetic Lung Cancer Therapy. Free Radic. Biol. Med. 2019, 130, 71–81. [Google Scholar] [CrossRef]
- Yazdani, Z.; Biparva, P.; Rafiei, A.; Kardan, M.; Hadavi, S. Combination Effect of Cold Atmospheric Plasma with Green Synthesized Zero-Valent Iron Nanoparticles in the Treatment of Melanoma Cancer Model. PLoS One 2022, 17, e0279120. [Google Scholar] [CrossRef]
- Qi, M.; Zhao, X.; Zhao, X.; Zhang, H.; Li, Z.; Zhang, X.; Fan, R.; Li, Q.; Zhang, J.; Xu, D. Violet Phosphorene Nanosheets and Cold Atmospheric Plasma for Synergetic Cancer Therapy. Chem. Eng. J. 2023, 145884. [Google Scholar] [CrossRef]
- Canady, J.; Murthy, S. R. K.; Zhuang, T.; Gitelis, S.; Nissan, A.; Ly, L.; Jones, O. Z.; Cheng, X.; Adileh, M.; Blank, A. T.; Colman, M. W.; Millikan, K.; O’Donoghue, C.; Stenson, K. M.; Ohara, K.; Schtrechman, G.; Keidar, M.; Basadonna, G. The First Cold Atmospheric Plasma Phase I Clinical Trial for the Treatment of Advanced Solid Tumors: A Novel Treatment Arm for Cancer. Cancers, 2023, 15, 3688. [Google Scholar] [CrossRef] [PubMed]
Figure 1.
A scatter chart comparing increase in the application of Machine Learning in Cancer and CAP, based on search results in PubMed & Google Scholar using two search strings, ("Machine Learning") AND (Cancer Treatment) & (“Machine Learning”) AND (“Cold Atmospheric Plasma”).
Figure 1.
A scatter chart comparing increase in the application of Machine Learning in Cancer and CAP, based on search results in PubMed & Google Scholar using two search strings, ("Machine Learning") AND (Cancer Treatment) & (“Machine Learning”) AND (“Cold Atmospheric Plasma”).

Figure 2.
Adaptive Plasma Framework for Precision Cancer Treatment: Integration of GP, MDP, Safe Q-Learning and Softmax[116].
Figure 2.
Adaptive Plasma Framework for Precision Cancer Treatment: Integration of GP, MDP, Safe Q-Learning and Softmax[116].
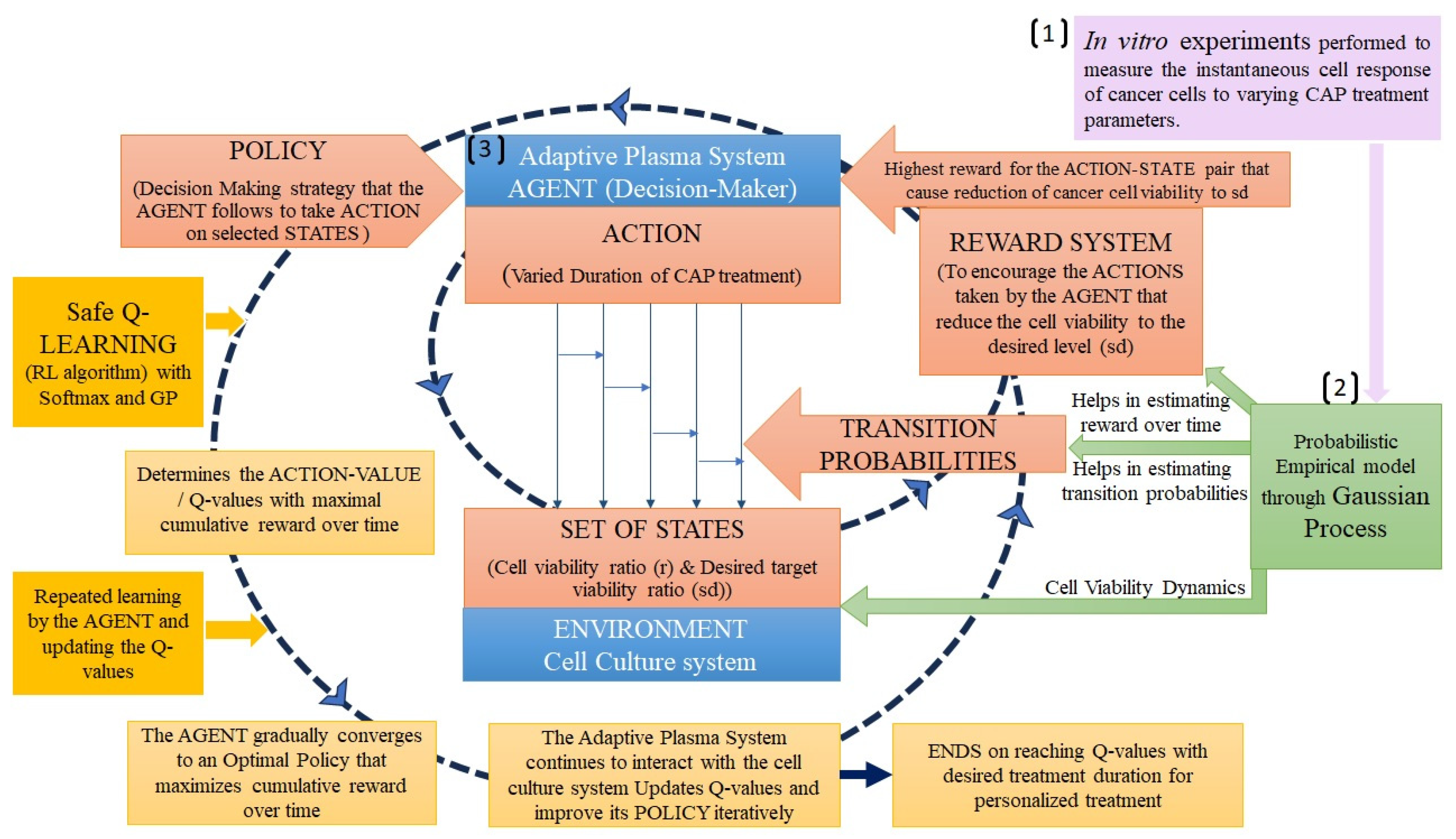
Figure 3.
Model Predictive Learning Control (MPLC) framework: Merging Real-Time data with Adaptive Plasma Control System to modify the treatment parameters online, making treatment more effective and personalized to each cancer cell’s response [122].
Figure 3.
Model Predictive Learning Control (MPLC) framework: Merging Real-Time data with Adaptive Plasma Control System to modify the treatment parameters online, making treatment more effective and personalized to each cancer cell’s response [122].

Figure 4.
Artificial Neural Networks (ANNs) in Real-Time diagnostics of plasma composition and Real-time control of plasma parameters to achieve optimized composition for personalized cancer treatment [123].
Figure 4.
Artificial Neural Networks (ANNs) in Real-Time diagnostics of plasma composition and Real-time control of plasma parameters to achieve optimized composition for personalized cancer treatment [123].

Figure 5.
Incorporating automatic real-time feedback control strategy into Adaptive Plasma system.
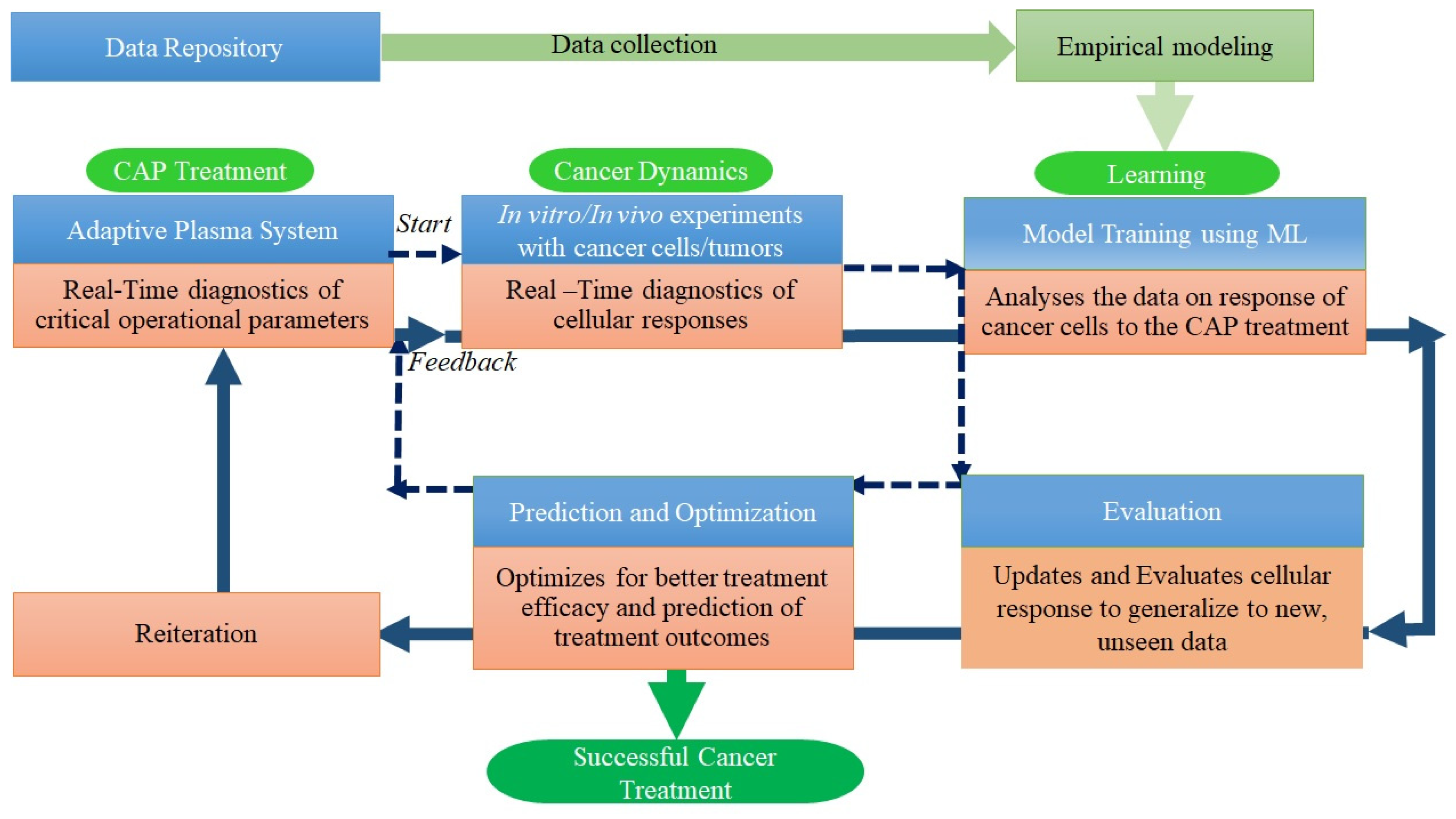
Table 1.
Selected parameters of the CAP sources for real-time diagnostics and AI techniques employed.
Table 1.
Selected parameters of the CAP sources for real-time diagnostics and AI techniques employed.
| Selected Parameters of the CAP sources for Real-Time diagnostics | Input Data obtained from | ML and computational techniques employed | Reference |
|---|---|---|---|
| Rotational and Vibrational temperatures | OES | Linear regression (Supervised ML) | [124] |
| Substrate characteristics | OES | k-Means Clustering (Unsupervised ML) | [124] |
| Separation distance between the electrodes | Electro-Acoustic Emission | Gaussian Process Regression (Supervised probabilistic ML) | [124] |
| Electron energy distribution function (EEDF) | OES | Genetic Algorithm (metaheuristic algorithm) | [129] |
| EEDF | OES, Momentum-transfer cross section | Visible Bremmsstrahlung Inversion (Supervised ML) | [130,131] |
| Time-series current signals from APPJ (discharge type and working gas) | Sensors/Probes | Convolutional neural networks (DL) | [132] |
| Plasma Plume length | Video frames of the plasma plume captured using a camera (iPhone 11) | Computer Vision algorithms | [133] |
| Temperature setpoint | Simulated data from thermal dynamics model of plasma-substrate interactions | Reinforcement learning | [127] |
| Self-Adaptive Plasma Chemistry Gas input densities and Energy levels |
OES | Artificial Neural Networks (DL), Gradual Mutation Algorithm | [119] |
| Pulse Discharge characteristics (current density and gap voltage) | Simulated fluid model data of time and pulse rise rate | Deep neural networks (DL) | [128] |
| Plasma chemistry (tokamak) | FTIR | Physics Informed Neural Networks | [134] |
Table 2.
Cancer cell response measured in Real-time in vitro studies.
| Input data | Real-time diagnostics | Advanced control and Prediction methodss | Reference |
|---|---|---|---|
| CAP treatment duration and Discharge voltage applied | Cell viability Luminescence Assay | Model Predictive Control (MPC) |
[67] |
| Cancer Cell viability ratio | Electrochemical Impedance Spectroscopy (EIS), operational parameters | GP regression, MPLC | [122] |
| Cancer Cell viability ratio | EIS, Cell viability assays, operational parameters | GP, Safety Q – Reinforcement learning | [116] |
| Voltage applied, irradiation time, frequency of the plasma and flow rate of the feed gas on the extent of DNA damage | Agarose gel electrophoresis, UV fluorescence Imaging | Artificial Neural Networks (supervised DL) Physics Guided Neural Network (supervised DL |
[136] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Copyright: This open access article is published under a Creative Commons CC BY 4.0 license, which permit the free download, distribution, and reuse, provided that the author and preprint are cited in any reuse.
MDPI Initiatives
Important Links
© 2024 MDPI (Basel, Switzerland) unless otherwise stated








